Chrysosplenium fallax Koldaeva (Chrysosplenium), a newly recorded species from China
-
Abstract:
Chrysosplenium fallaxKoldaeva, recently discovered and collected in Yanji, Jilin Province, represents a newly recorded species of Saxifragaceae in China. This species, native to the Russian Far East, was first described as a new species in 2021. Based on precise field investigations and specimen examination, we provide a comprehensive description ofC. fallaxand its seed micromorphology. Phylogenetic analysis based on chloroplast genomes of 45Chrysospleniumspecies confirmed the systematic position ofC. fallaxinChrysosplenium. Voucher specimens were deposited in the Herbarium of South-Central Minzu University (HSN).
-
Keywords:
- Chrysosplenium fallax /
- Chrysosplenium /
- Newly record /
- Phylogeny /
- China
-
Chrysosplenium is a genus of small perennial herbaceous plants widely distributed in the Northern Hemisphere, holding a prominent taxonomic position within Saxifragaceae phylogeny[1-3]. Chrysosplenium is readily distinguished from other related genera, such as Bergenia, Saxifraga, and Tiarella, by its tetramerous flowers and petaloid sepals[4]. Chrysosplenium species contain highly hydroxylated, methoxylated flavonoids and triterpenoids with significant medicinal value, with some species also used ornamentally[5, 6]. To date, more than 80 species of Chrysosplenium have been documented, including 40 species in China[7, 8], 24 (60%) of which are endemic to China, primarily found in the Shaanxi, Sichuan, and Yunnan provinces[9-11].
During a field investigation in Yanji, northeast China, we collected a distinctive species of Chrysosplenium. While morphologically similar to C. pilosum Maxim., the specimens were distinguished by the presence of four large, accumulated leaves at the top of sterile branches. Extensive literature review, specimen examination, and material observations[8-16] confirmed that the Yanji specimens belong to C. fallax Koldaeva, a species recently published by Russian scholars in 2021[10]. C. fallax has a previously reported distribution restricted to the Russian Far East, including the Shkotovsky District, Ussuriysky and Vladivostoksky (except islands) UrbanOkrugs. Therefore, our identification of C. fallax as a newly recorded species in Yanji, China, is significant. In this study, we provide a detailed morphological description of C. fallax, along with accompanying color photographs. Furthermore, we performed a phylogenetic analysis of C. fallax based on 78 chloroplast coding sequences. Voucher specimens were deposited in the Herbarium of South-Central Minzu University (HSN).
1. Materials and Methods
1.1 Morphological observations
The morphological characteristics of plant material from field and herbarium specimens were studied using a dissecting microscope (SMZ171, Motic, China). A scanning electron microscope(SEM) was employed to observe seed morphology. Seed materials were collected from the field and dried with silica gel. Pre-treatment of seeds followed previous research[12]. A Hitachi SU8010 SEM was used for observations and photography. Seed size and surface were determined using at least 15 seeds.
1.2 Phylogenetic analysis
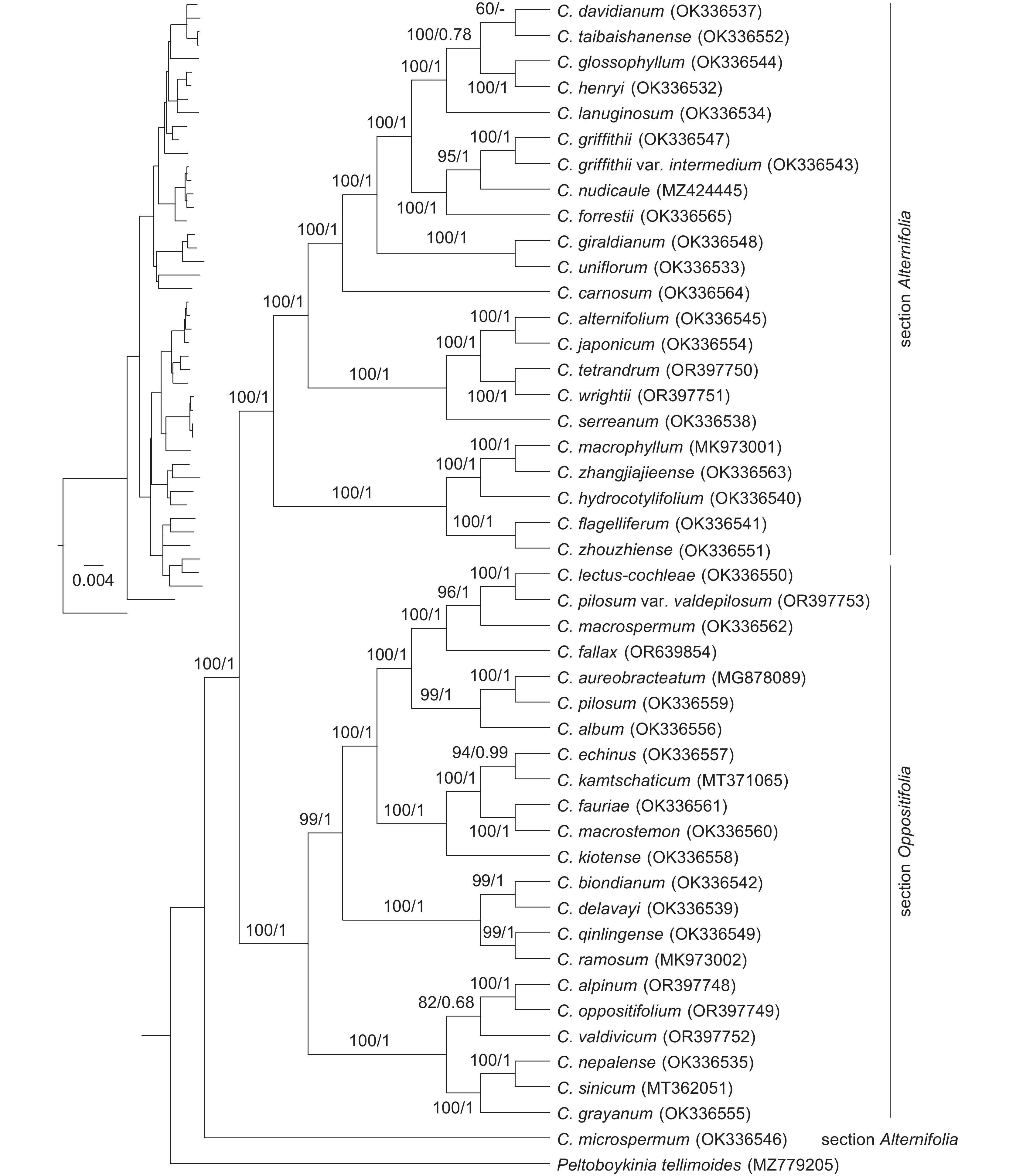
To confirm the placement of C. fallax within Chrysosplenium, phylogenetic analysis was conducted using chloroplast protein-coding genes. Analysis included 45 species of Chrysosplenium as the in-group and Peltoboykinia tellimoides (Maxim.) H. Hara as the out-group. The chloroplast genome of C. fallax was newly generated in this study, while the sequences of the other species were obtained from GenBank (https://www.ncbi.nlm.nih.gov/). The GenBank accession numbers for all species are listed in Table 1.
Table 1. Species names and GenBank accession numbers used in this studyNumber Taxon Section Chloroplast genome 1 Chrysosplenium album Maxim. Oppositifolia OK336556 2 Chrysosplenium alpinum Schur Oppositifolia OR397748 3 Chrysosplenium aureobracteatum Y. I. Kim & Y. D. Kim Oppositifolia MG878089 4 Chrysosplenium biondianum Engl. Oppositifolia OK336542 5 Chrysosplenium delavayi Franch. Oppositifolia OK336539 6 Chrysosplenium echinus Maxim. Oppositifolia OK336557 7 Chrysosplenium fallax Koldaeva Oppositifolia OR639854 8 Chrysosplenium fauriae Franch. Oppositifolia OK336561 9 Chrysosplenium grayanum Maxim. Oppositifolia OK336555 10 Chrysosplenium kamtschaticum Fisch. ex Seringe Oppositifolia MT371065 11 Chrysosplenium kiotense Ohwi. Oppositifolia OK336558 12 Chrysosplenium lectus-cochleae Kitagawa Oppositifolia OK336550 13 Chrysosplenium macrospermum Y. I. Kim & Y. D. Kim Oppositifolia OK336562 14 Chrysosplenium macrostemon Maxim. ex Franch. et Sav. Oppositifolia OK336560 15 Chrysosplenium nepalense D. Don Oppositifolia OK336535 16 Chrysosplenium oppositifolium L. Oppositifolia OR397749 17 Chrysosplenium pilosum Maxim. Oppositifolia OK336559 18 Chrysosplenium pilosum Maxim. var. valdepilosum Oppositifolia OR397753 19 Chrysosplenium qinlingense Z. P. Jien ex J. T. Pan Oppositifolia OK336549 20 Chrysosplenium ramosum Maxim. Oppositifolia MK973002 21 Chrysosplenium sinicum Maxim. Oppositifolia MK814606 22 Chrysosplenium valdivicum Hook. Oppositifolia OR397752 23 Chrysosplenium alternifolium L. Alternifolia OK336545 24 Chrysosplenium carnosum Hook. f. et Thoms. Alternifolia OK336564 25 Chrysosplenium davidianum Decne. ex Maxim. Alternifolia OK336537 26 Chrysosplenium flagelliferum Fr. Schmidt. Alternifolia OK336541 27 Chrysosplenium forrestii Diels Alternifolia OK336565 28 Chrysosplenium giraldianum Engl. Alternifolia OK336548 29 Chrysosplenium glossophyllum Hara Alternifolia OK336544 30 Chrysosplenium griffithii Hook. f. et Thoms. Alternifolia OK336547 31 Chrysosplenium griffithii var. intermedium (Hara) J. T. Pan Alternifolia OK336543 32 Chrysosplenium henryi Franch. Alternifolia OK336532 33 Chrysosplenium hydrocotylifolium Lévl. et Vant. Alternifolia OK336540 34 Chrysosplenium japonicum (Maxim.) Makino Alternifolia OK336554 35 Chrysosplenium lanuginosum Hook. f. et Thoms. Alternifolia OK336534 36 Chrysosplenium macrophyllum Oliv. Alternifolia MK973001 37 Chrysosplenium microspermum Franch. Alternifolia OK336546 38 Chrysosplenium nudicaule Bunge Alternifolia MZ424445 39 Chrysosplenium serreanum Hand.-Mazz. Alternifolia OK336538 40 Chrysosplenium taibaishanense J. T. Pan Alternifolia OK336552 41 Chrysosplenium tetrandrum (N. Lund) Th. Fries Alternifolia OR397750 42 Chrysosplenium uniflorum Maxim. Alternifolia OK336533 43 Chrysosplenium wrightii Franch. & Sav. Alternifolia OR397751 44 Chrysosplenium zhangjiajieense X. L. Yu, Hui Zhou & D. S. Zhou Alternifolia OK336563 45 Chrysosplenium zhouzhiense Hong Liu Alternifolia OK336551 46 Peltoboykinia tellimoides (Maxim.) Hara — MZ779205 PhyloSuite v1.2.2[17] was used to extract 78 protein-coding genes from the chloroplast genome. The coding sequences were aligned using MAFFT v7.4[18], then combined using PhyloSuite v1.2.2[17]. Maximum-likelihood (ML) analysis was performed using IQ-TREE v2.1.2[19] with 1 000 bootstrap replicates. The best-fit substitution model (TVM+F+R4) was determined using ModelFinder[20]. The Bayesian inference (BI) tree was obtained using MrBayes v3.2.6[21], run for one million generations, one run, and two chains, with 25% of trees discarded as burn-in and trees sampled every 1 000 generations (1 000 trees sampled in total) using the GTR model. FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/fgtree/) was used for tree visualization.
2. Taxonomic Treatment
宽卵叶金腰(新拟,Fig. 1)
Chrysosplenium fallax Koldaeva, Phytotaxa 491(1): 039, Fig. 1-4, 2021.
Type: Russian Federation, Primorsky Krai, Vladivostoksky Urban Okrug, Muravyev-Amursky Peninsula, Malaya Pionerskaya River valley, edge of a coniferous-broad-leaf forest, moist habitat, 12 May 2017, M.N. Koldaeva 0115.
Perennial herbs, 10–20 cm high, hermaphroditic, pulpy, pubescent with long, white hairs, reddish when dry. Root fibrous and soft, 1.2–5 cm long, underground, plagiotropic, with 2–5 elongated internodes. Flowering stem single, erect, 14–20(–24) cm tall, light green to green, tetragonal in cross-section, middle part sparsely pubescent, base and apical part almost glabrous, with 2–7 sterile branches arising from all leaf axils. Vegetative shoots above-ground and underground. Underground stolons 1–4, 3 to 48 cm long, 1.5–2 mm in diameter, unbranched or weakly branched in distal part, white; Approximately 10-cm-long distal portion of stolon remains by end of growing season. Repent stolon-like shoots 3–8, evenly leafed; leaves 5–6 pairs, similar, small. Sterile branches orthotropic (axial and lateral), prostrate, repent, stolon-like, pubescent with long hairs, most densely aggregated at apical part; Axial sterile branches 6 to 21 cm long, with branches or non-branching; Lateral sterile branches 5 to 16 cm long, with basal internode elongated. Leaves simple, estipulate, petiolate. Basal leaves absent. Cauline leaves 2–6, opposite or rarely alternate, petioles 3–9 mm long, sparsely pilose; blade 3–13×4–19 mm, subflabellate or flabellate-rounded, apex subtruncate to rounded, base cuneate to broadly cuneate, adaxial surface glabrous, abaxial surface glabrous or with single hairs along veins at base, often with red specks, margins ciliate, with 5–7–9 smooth-serrated crenates. Leaves of sterile branches opposite, not evenly leafed, highly unequal, with largest ones acervate, resembling rosette at apex but not forming true rosette, apical part not rooted; leaves of axial sterile branches 4–9(–11) pairs; blade 2.1–5.6×2.0–3.7 cm (except uppermost one), distal 2–3 pairs largest, with short internodes (0.2–0.5 cm); blade rounded, rounded-ovate, or elliptical; base cuneate; margin undulate-crenate, crenate-sinuate, or almost smooth-ciliate, with 5–7 crenates on each side; leaves of lateral sterile branches (1–)2–4 pairs, acervate to resemble rosette at apex; pubescence of leaf abaxial side along main veins on 1st or 2nd pairs of lateral branches or 2nd to 5th pairs of axial branches; in upper pairs, pubescence becomes denser; blade adaxial side glabrous or with solitary hairs at periphery, sometimes with silvery dots; petioles sparsely pilose on both sides. Cymes compact, 5–18 flowered, surrounded by large, greenish-yellow bracts. Inflorescence branches and bracteal petioles with solitary long hairs. Bracteal leaves greenish-yellow, petioles 1–4 mm, pilose; blades 4–16×2–12 mm, flabelliform or broadly ovate to ovate, apex subtruncate to rounded, base cuneate to broadly cuneate, with 5–7–9 smoothed-serrated crenates, upper surface glabrous, densely silvery dotted, lower surface glabrous, greenish-gray. Flowers tetramerous, actinomorphic, goblet-shaped, yellow, sometimes with red specks, pedicels short (0.8–3 mm). Sepals 4, erect, convex outside, with tip slightly bent outward; external almost rounded 2.2×2.1 mm; internal ovoid 1.8×1.5 mm, shorter than external ones. Petals absent. Stamens 8, biseriate, ca. 2 mm long, equal to calyx, filaments filiform, 1.5 mm long. Anthers yellow, 2-locular, ca. 0.5 mm long, longitudinally dehiscent. Pistil 2-carpellate, subsuperior, ovary 1-locular, ovules at two parietal placentae, styles 2, erect, slightly divergent, ca. 1 mm long, stigma round, disc absent; nectary not prominent, green. Capsule 6–8 mm, light green, glabrous, ca. 2–2.5 times longer than calyx, slightly unequal, divergent, arcuately curved. Seeds numerous, brown, suborbicular, minutely papillose, with 18–23 longitudinal rows of large, thick-walled, 0.7–0.8×0.6–0.7 mm, densely located tubercles. Tubercles almost cylindrical, with one, rarely two papillae.
Phenology: The flowering period of C. fallax occurs from early to late May and the fruiting period occurs from early to late June.
Specimens examined: China, Jilin Province, Yanji City, Yanbian County, 42°59′12″N, 129°16′50″E, 300 m, 29 May 2021, H. Liu et X. T. Chen
20210529001 (HSN).Distribution and habitat: The species C. fallax is currently known from the Russian Far East and China(Jilin). In China, it has only been observed in limited areas of Yanji. The species grows in broad-leaf forests in moist and damp areas on gentle slopes along river valleys and streams, with an elevational range of 20–380 m.
Phylogenetic position: Phylogenetic analyses indicated that C. fallax belongs to Chrysosplenium sect. Oppositifolia and forms a strongly supported clade with C. macrospermum Y. l. Kim & Y. D. Kim, C. pilosum Maxim. var. valdepilosum, C. lectus-cochleae kitagawe, C. aureobracteatum Y. l. Kim & Y. D. Kim, C. pilosum Maxim., and C. album Maxim. (Fig. 2). These species, which belong to ser. Pilosa Maxim., are primarily distributed in northeastern China, Russian Far East, Japan, and North and South Korea.
Discussion
Phylogenetic trees were constructed for 45 species of Chrysosplenium using the ML and BI approaches. The resulting topologies from both methods were similar, confirming the monophyletic nature of the genus Chrysosplenium, as supported by previous research[2, 3, 22]. Our results further revealed that Chrysosplenium could be divided into three major clades, with C. microspermum positioned at the base of the phylogenetic tree, consistent with prior findings[22]. Phylogenetic analysis also demonstrated that C. fallax belongs to Chrysosplenium sect. Oppositifolia and clusters with six other species within ser. Pilosa, which contains around 20 species[8, 14]. Morphologically, C. fallax is characterized by erect sepals, opposite leaves, and pilose stems, which align with the features of ser. Pilosa[8, 14]. Geographically, C. fallax has been found in the Russian Far East and Jilin Province in northeast China, corresponding to the primary distribution of ser. Pilosa in northeast Asia[8, 14]. Although C. fallax and C. pilosum are not phylogenetically closely related, they share significant morphological similarities. Notably, C. fallax can be distinguished from C. pilosum by vegetative shoots lacking branching in distal part, with larger acervate leaves on top, pubescent on abaxial side of leaf blade. Additionally, C. fallax is morphologically similar to C. villosum, a species not included in our phylogenetic tree, but can be distinguished based on plagiotropic rhizome with long internodes and specialized underground stolons. Currently, five species in ser. Pilosa have been discovered in China, including C. fallax, C. pilosum, C. macrospermum, C. hebetatum, and C. lectus-cochleae. The discovery of C. fallax has expanded the distribution range of ser. Pilosa and enriched the background data on Chrysosplenium in China.
-
Table 1 Species names and GenBank accession numbers used in this study
Number Taxon Section Chloroplast genome 1 Chrysosplenium album Maxim. Oppositifolia OK336556 2 Chrysosplenium alpinum Schur Oppositifolia OR397748 3 Chrysosplenium aureobracteatum Y. I. Kim & Y. D. Kim Oppositifolia MG878089 4 Chrysosplenium biondianum Engl. Oppositifolia OK336542 5 Chrysosplenium delavayi Franch. Oppositifolia OK336539 6 Chrysosplenium echinus Maxim. Oppositifolia OK336557 7 Chrysosplenium fallax Koldaeva Oppositifolia OR639854 8 Chrysosplenium fauriae Franch. Oppositifolia OK336561 9 Chrysosplenium grayanum Maxim. Oppositifolia OK336555 10 Chrysosplenium kamtschaticum Fisch. ex Seringe Oppositifolia MT371065 11 Chrysosplenium kiotense Ohwi. Oppositifolia OK336558 12 Chrysosplenium lectus-cochleae Kitagawa Oppositifolia OK336550 13 Chrysosplenium macrospermum Y. I. Kim & Y. D. Kim Oppositifolia OK336562 14 Chrysosplenium macrostemon Maxim. ex Franch. et Sav. Oppositifolia OK336560 15 Chrysosplenium nepalense D. Don Oppositifolia OK336535 16 Chrysosplenium oppositifolium L. Oppositifolia OR397749 17 Chrysosplenium pilosum Maxim. Oppositifolia OK336559 18 Chrysosplenium pilosum Maxim. var. valdepilosum Oppositifolia OR397753 19 Chrysosplenium qinlingense Z. P. Jien ex J. T. Pan Oppositifolia OK336549 20 Chrysosplenium ramosum Maxim. Oppositifolia MK973002 21 Chrysosplenium sinicum Maxim. Oppositifolia MK814606 22 Chrysosplenium valdivicum Hook. Oppositifolia OR397752 23 Chrysosplenium alternifolium L. Alternifolia OK336545 24 Chrysosplenium carnosum Hook. f. et Thoms. Alternifolia OK336564 25 Chrysosplenium davidianum Decne. ex Maxim. Alternifolia OK336537 26 Chrysosplenium flagelliferum Fr. Schmidt. Alternifolia OK336541 27 Chrysosplenium forrestii Diels Alternifolia OK336565 28 Chrysosplenium giraldianum Engl. Alternifolia OK336548 29 Chrysosplenium glossophyllum Hara Alternifolia OK336544 30 Chrysosplenium griffithii Hook. f. et Thoms. Alternifolia OK336547 31 Chrysosplenium griffithii var. intermedium (Hara) J. T. Pan Alternifolia OK336543 32 Chrysosplenium henryi Franch. Alternifolia OK336532 33 Chrysosplenium hydrocotylifolium Lévl. et Vant. Alternifolia OK336540 34 Chrysosplenium japonicum (Maxim.) Makino Alternifolia OK336554 35 Chrysosplenium lanuginosum Hook. f. et Thoms. Alternifolia OK336534 36 Chrysosplenium macrophyllum Oliv. Alternifolia MK973001 37 Chrysosplenium microspermum Franch. Alternifolia OK336546 38 Chrysosplenium nudicaule Bunge Alternifolia MZ424445 39 Chrysosplenium serreanum Hand.-Mazz. Alternifolia OK336538 40 Chrysosplenium taibaishanense J. T. Pan Alternifolia OK336552 41 Chrysosplenium tetrandrum (N. Lund) Th. Fries Alternifolia OR397750 42 Chrysosplenium uniflorum Maxim. Alternifolia OK336533 43 Chrysosplenium wrightii Franch. & Sav. Alternifolia OR397751 44 Chrysosplenium zhangjiajieense X. L. Yu, Hui Zhou & D. S. Zhou Alternifolia OK336563 45 Chrysosplenium zhouzhiense Hong Liu Alternifolia OK336551 46 Peltoboykinia tellimoides (Maxim.) Hara — MZ779205 -
[1] Soltis DE,Morgan DR,Grable A,Soltis PS,Kuzoff R. Molecular systematics of Saxifragaceae sensu stricto[J]. Am J Bot,1993,80(9):1056−1081. doi: 10.1002/j.1537-2197.1993.tb15333.x
[2] Soltis DE,Soltis PS. Phylogenetic relationships in Saxifragaceae sensu lato:a comparison of topologies based on 18S rDNA and rbcL sequences[J]. Am J Bot,1997,84(4):504−522. doi: 10.2307/2446027
[3] Deng JB,Drew BT,Mavrodiev EV,Gitzendanner MA,Soltis PS,Soltis DE. Phylogeny,divergence times,and historical biogeography of the angiosperm family Saxifragaceae[J]. Mol Phylogenet Evol,2015,83:86−98. doi: 10.1016/j.ympev.2014.11.011
[4] Kim YI,Shin JS,Lee S,Chen JH,Choi S,et al. A new species of Chrysosplenium (Saxifragaceae) from Northeastern China[J]. PhytoKeys,2019,135(3):39−47.
[5] Zhao JJ,Qiu X,Zhao YY,Wu R,Wei PH,et al. A review of the genus Chrysosplenium as a traditional Tibetan medicine and its preparations[J]. J Ethnopharmacol,2022,290:115042. doi: 10.1016/j.jep.2022.115042
[6] Pan JT. A study on the genus Chrysosplenium L. from China[J]. Acta Phytotaxon Sin,1986,24(2):81−97.
[7] Xiang NY,Lu BJ,Yuan T,Yang TG,Guo JN,et al. De novo transcriptome assembly and EST-SSR marker development and application in Chrysosplenium macrophyllum[J]. Genes,2023,14(2):279. doi: 10.3390/genes14020279
[8] Hara H. Synopsis of the genus Chrysosplenium L. (Saxifragaceae)[J]. J Faculty Sci Univ Tokyo,Sect 3,Bot,1957,7:1−90.
[9] Fu LF,Yang TG,Lan DQ,Wen F,Liu H. Chrysosplenium sangzhiense (Saxifragaceae),a new species from Hunan,China[J]. PhytoKeys,2021,176:21−32. doi: 10.3897/phytokeys.176.62802
[10] Koldaeva MN. Chrysosplenium fallax (Saxifragaceae),a new species from the Russian Far East[J]. Phytotaxa,2021,491(1):35−46. doi: 10.11646/phytotaxa.491.1.4
[11] Pan JT,Ohba H. Chrysosplenium[M]//Wu ZY,Raven PH,eds. Flora of China:Vol. 8. Beijing:Science Press,2001:346−358.
[12] Fu LF,Liao R,Lan DQ,Wen F,Liu H. A new species of Chrysosplenium (Saxifragaceae) from Shaanxi,north-western China[J]. PhytoKeys,2020,159:127−135. doi: 10.3897/phytokeys.159.56109
[13] Liu H,Luo JL,Liu QY,Lan DQ,Qin R,Yu XL. A new species of Chrysosplenium (Saxifragaceae) from Zhangjiajie,Hunan,central China[J]. Phytotaxa,2016,277(3):287−292. doi: 10.11646/phytotaxa.277.3.7
[14] Kim YI,Cho SH,Lee JH,Kang DH,Park JH,Kim YD. Chrysosplenium ramosissimum Y. I . Kim & Y. D. Kim (Saxifragaceae),a new species from Korea[J]. PhytoKeys,2018,111:1−10.
[15] Kim YI,Kim YD. Chrysosplenium aureobracteatum(Saxifragaceae),a new species from south Korea[J]. Novon:A Journal for Botanical Nomenclature,2015,23(4):432−436. doi: 10.3417/2013018
[16] Han JW,Kang SH. Chrysosplenium epigealum J. W. Han et S. H. Kang:a new species of Chrysosplenium(Saxifragaceae) from Korea[J]. Korean J Plant Res,2012,25(3):346−348. doi: 10.7732/kjpr.2012.25.3.346
[17] Zhang D,Gao FL,Jakovlić I,Zou H,Zhang J,et al. PhyloSuite:an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies[J]. Mol Ecol Resour,2020,20(1):348−355. doi: 10.1111/1755-0998.13096
[18] Katoh K,Standley DM. MAFFT multiple sequence alignment software version 7:improvements in performance and usability[J]. Mol Biol Evol,2013,30(4):772−780. doi: 10.1093/molbev/mst010
[19] Nguyen LT,Schmidt HA,Von Haeseler A,Minh BQ. IQ-TREE:a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies[J]. Mol Biol Evol,2015,32(1):268−274. doi: 10.1093/molbev/msu300
[20] Kalyaanamoorthy S,Minh BQ,Wong TKF,von Haeseler A,Jermiin LS. ModelFinder:fast model selection for accurate phylogenetic estimates[J]. Nat Methods,2017,14(6):587−589. doi: 10.1038/nmeth.4285
[21] Ronquist F,Teslenko M,van der Mark P,Ayres DL,Darling A,et al. MrBayes 3.2:efficient Bayesian phylogenetic inference and model choice across a large model space[J]. Syst Biol,2012,61(3):539−542. doi: 10.1093/sysbio/sys029
[22] Yang TG,Wu ZH,Tie J,Qin R,Wang J,Liu HQ. A comprehensive analysis of chloroplast genome provides new insights into the evolution of the genus Chrysosplenium[J]. Int J Mol Sci,2023,24(19):14735. doi: 10.3390/ijms241914735



 下载:
下载: