Research progress on the genetic regulatory mechanism of flowering in Chrysanthemum
-
摘要:
开花是植物发育过程中一个关键的质变过程,是植物从营养生长向生殖生长阶段的转变。对于观赏植物来说,开花的早晚决定了其市场应用和经济价值。植物开花受到内外信号的复杂调控,基于模式植物拟南芥(Arabidopsis thaliana (L.) Heynh)的研究,目前已经阐明了6条主要的开花调控途径,这些途径彼此独立又互相交叉,形成复杂的遗传调控网络。菊花(Chrysanthemum × morifolium Ramat)作为起源于中国的世界名花,是世界花卉市场的重要一员,但因其是典型的短日照植物,不仅增加了生产中开花期调控成本,也限制了菊花的应用范围。本文以高等植物开花遗传调控网络为基础,综述了菊花开花遗传调控机制的研究进展,以期为菊花开花时间改良育种工作提供理论指导,同时也为解析高等植物开花机制提供新见解。
Abstract:Flowering represents a critical transition in plant development, shifting from the vegetative to reproductive growth stages. In ornamental plants, the timing of flowering significantly impacts marketability and economic value. Plant flowering is regulated by complex internal and external signals. Studies on the model plant Arabidopsis thaliana have identified six primary pathways related to flowering regulation. These independent but intersecting pathways form a complex genetic regulatory network. Chrysanthemum × morifolium, a famous flower originating from China, holds a considerable share of the world flower market. However, its typical short-day flowering requirements not only increase production costs but also limit its application scope. Based on the flowering regulatory networks of higher plants, this review discusses current research progress on the genetic regulatory mechanisms underlying chrysanthemum flowering, thus providing theoretical guidance for the breeding and improvement of flowering time, as well as new insights into the flowering mechanisms of higher plants.
-
Keywords:
- Chrysanthemum × morifolium /
- Flowering /
- Environmental factors /
- Genetic regulation
-
TRANSPARENT TESTA GLABRA1(TTG1)是一种串连肽重复蛋白[1],含有7个WD40结构域。TTG1基因对植物生长发育具有重要意义,能够参与调控多种生命活动,譬如:调节花青素的积累[2]、表皮毛和根毛的密度[3],影响种子大小、种皮颜色以及种皮粘液的合成[4],调控开花时间[5]等。
花青素作为类黄酮化合物[6],具有较强的抗氧化[7]和抗炎[8]作用。植物中花青素的积累,使植物具有吸引传粉者[9]、免受紫外线或病虫侵害[10]等多种生物学功能。TTG1在花青素合成过程中极为重要,突变体ttg1中花青素的积累受阻[11]。拟南芥(Arabidopsis thaliana (L.) Heynh.)中,花青素转录激活有两种调控方式,一种是不依赖bHLH转录因子的MYB转录因子和WD40蛋白形成的二元复合体协同调控,如:TTG1-GL3/TT8-TT2复合物通过激活DFR、ANS、BAN、TT9和TT12的表达来调控花青素的生物合成[12-14];另一种是MYB转录因子、bHLH转录因子以及WD40蛋白形成三元复合体来协同调控,如:TTG1-TT8/GL3-PAP1/PAP2/MYB113/MYB114复合物通过激活DFR、ANS、BAN、UF3GT的表达来调控花青素的生物合成[15-17]。
表皮毛是植物地上部分表面的附属物,具有减少水分蒸发及病虫侵害的作用[18]。研究表明,R2R3-MYB型转录因子(MYB23、MYB82、GL3)、MYC1编码的bHLH型转录因子[19]、WD40类重复蛋白(TTG1、TT8、GL3、EGL3)[20, 21]、C2H2锌指蛋白家族基因(GIS、GIS2、GIS3、ZFP5、ZFP6、ZFP8)等能够正调控拟南芥表皮毛的发育[22-24];而R3-MYB转录因子(ETC1、ETC2、ETC3、TCL1、TCL2、TRY、CPC等)能够负调控其发育[25]。突变体ttg1具有无表皮毛的特征[11],说明TTG1在表皮毛发育过程中起着关键作用。TTG1能够与R2R3-MYB转录因子中的GL1、MYB23以及bHLH转录因子中的GL3、EGL3相互作用,形成MBW,激活下游基因GL2的表达,从而促进表皮毛的生长发育[26]。TTG1也能够与SPL转录因子中的SPL4或SPL5相互作用,从而影响MBW复合物的转录活性,导致毛状体起始受到抑制[27]。
开花是植物从营养期向生殖期过渡的重要性状。研究发现,ttg1突变体比野生型早开花,TTG1过表达植株则比野生型晚开花[5],表明TTG1能够调控植物的开花时间。FLC是编码MADS-box转录因子的开花抑制基因[28],TTG1位于FLC的上游,Paffendorf 等[5]发现TTG1可以激活FLC的表达,调节开花时间。FT是一种成花素基因[29],通常在叶片中表达,转移到茎尖分生组织,进而诱导开花[14]。FLC通过与FT的启动子区域相结合,可以抑制FT的表达,促使植物形成晚花表型。
TTG1除了能够影响拟南芥的花青素生物合成、表皮毛生长以及开花期外,还能影响其种皮粘液的产生以及根毛密度等[11]。TTG1对种皮粘液的调控也是依靠形成MBW复合物来进行,MBW复合物可以激活TTG2和GL2的表达,进而激活粘液生物合成基因MUM4的表达,导致种皮粘液的产生[30]。TTG1除了可以直接调节粘液生物合成基因的表达,还可以通过调节种子中脂肪酸的积累来间接调节种皮粘液的产生[31]。TTG1参与形成的MBW复合物能够调节植物激素乙烯(ET)的形成[32],进而影响植物根毛密度。
前期研究中,我们从水稻(Oryza sativa L.)中图位克隆了OsTTG1基因,并利用CRISPR/Cas9技术研究了其功能,发现OsTTG1可以调控水稻叶片、叶鞘和种皮等组织部位的花青素生物合成[7]。本研究利用转基因技术在拟南芥突变体ttg1中过表达OsTTG1基因,研究结果旨在探究该基因的其他生物学功能,并为其开发利用提供科学依据。
1. 材料与方法
1.1 材料
拟南芥突变体ttg1购自Arashare(中国拟南芥突变体服务中心)。野生型Col-0和OsTTG1过表达株系OsTTG1-OE由本实验保存。
1.2 材料种植和表型观察
取适量拟南芥种子放入1.5 mL的离心管中,向离心管加入0.1%的琼脂糖溶液,并将其放入4 ℃冰箱中保存3~4 d[33]。将春化处理过的种子均匀地播撒于基质中,在温度为18 ℃~20 ℃、湿度为60%~70%的培养箱中培养。
分别取拟南芥Col-0、ttg1、OsTTG1-OE的种子、植株从根基部开始往上1 cm左右的茎段、以及叶片和花序等材料,放置到体视显微镜下观察,并拍照保存。使用佳能EOS R5相机对莲座期、开花期、结荚期的拟南芥进行拍照。不同基因型的拟南芥在相同条件、相同设置下进行观察及拍照。
1.3 OsTTG1基因的克隆与载体的构建
通过PCR方法克隆目的基因,用琼脂糖凝胶电泳确定目的条带的位置,按照胶回收试剂盒说明书,进行目的片段的回收。PCR引物为CAM-FLAG-
45810 (F:5'-TTTGGAGAGGACACGCTCGAGATGGAGCAGCCCAAGCCG-3';R:5'-GTCATCCTTGTAGTCGAATTCGACCCTGAGAAGCTGGACCTTGT-3')。用EocRⅠ和XhoⅠ对质粒CAM-flag进行双酶切,琼脂糖凝胶电泳检测酶切产物,回收载体大片段对应的条带。将目的DNA片段与线性化载体进行连接,形成重组克隆载体。将连接产物转化大肠杆菌感受态细胞DH5α,使用单菌落PCR进行阳性克隆鉴定。利用Plasmid Mini Kit Ⅰ试剂盒提取测序正确的质粒,采用冻融法直接转化农杆菌GV3101,使用单菌落PCR进行阳性克隆鉴定。单菌落PCR检测的引物均为P(F:5'-AGAAGACGTTCCAACCACG-3';R:5'-CGGTAAGGATCTGAGCTACAC-3')。1.4 拟南芥转基因及转化植株的筛选
将含阳性重组质粒的农杆菌接种于含有抗生素的LB液体培养基中,获得转化液。采用花序侵染法转化拟南芥[34],收集T0代种子。将T0代种子播种于含有抗生素的MS培养基中,筛选具有抗性的拟南芥植株。选取生长健壮的植株移栽至育苗基质中进行培养。待种子成熟后,按单株收集T1代种子,连续培养具有抗性的拟南芥,获得纯合的T3代种子,用于后续实验以及表型鉴定。
1.5 转基因植株PCR鉴定及RT-PCR分析
提取拟南芥野生型和转基因阳性植株的DNA。分别以转基因、野生型植株的DNA、H2O和农杆菌菌液为模板,进行PCR鉴定,引物为45810(F:5'-CTTCACCTGCGAGCTGC-3';R:5'-CTTGTCGTGCGCGATGAG-3')。
对拟南芥野生型、突变体ttg1和OsTTG1-OE株系进行RT-PCR分析。总RNA提取按照 RNeasy Plant Mini kit 说明书进行。使用TaKaRa试剂盒进行反转录,获得cDNA。以ACTIN2为内参,利用RT-PCR检测AtTTG1和OsTTTG1在不同基因型中的表达情况。引物分别为:ACTIN2(F:5'-ACCTTGCTGGACGTGACCTTACTGAT-3';R:5'-GTTGTCTCGTGGATTCCAGCAGCTT-3');TTG1(F:5'-CAACATATGATGGATAATTCAGCTCCAGATTCG-3';R:5'-CAACTTAAGTCAAACTCTAAGGAGCTGCATTTTG-3')以及OsTTG1(F:5'-TTACTCTGAACTCCCCGACCCG-3';R:5'-GTTGGATTCGCTTCGCTTTGCT-3')。
1.6 RNA-seq分析
为进一步挖掘引起OsTTG1-OE表型变化的基因,选取莲座期、抽薹期和结荚期3个关键时期,对ttg1和OsTTG1-OE植株的地上部分进行取样,每个时期、每种基因型取4个重复,共24个样品,进行转录组测序。
提取24个样品的总RNA,利用Nanodrop 2000对RNA的浓度和纯度进行检测,用琼脂糖凝胶电泳检测RNA的完整性,使用Agilent 2100测定RIN值。将检测合格的样品进行文库构建,确保文库质量合格,然后选择Illumina平台,进行上机测序。
1.7 生物信息学分析流程
使用TopHat 2软件将得到的Clean data与指定的参考基因组进行序列比对,获得Mapped data。参考基因组版本为TAIR10,来源于网站http://plants.ensembl.org/Arabidopsis_thaliana/Info/Index,同时对测序的结果进行质量评估。使用DESeq 2软件进行差异表达基因(Differentially expressed genes, DEGs)分析,筛选阈值为:|log2FC|≥1,且 padjust<0.05。结合GraphPad Prism 5.0软件和在线网站(https://www.bioinformatics.com.cn/)等进行作图分析。
2. 结果与分析
2.1 植物表达载体的构建和遗传转化拟南芥
用EocRⅠ和XhoⅠ双酶切质粒CAM-flag,回收载体大片段。将目的DNA片段与线性化载体连接,连接产物转化大肠杆菌感受态细胞DH5α,获得了阳性克隆。
利用试剂盒提取测序正确的重组质粒,采用冻融法直接转化农杆菌GV3101。经抗性平板筛选,菌落PCR进行阳性克隆鉴定,发现可扩增出明亮单一的条带,且与预期片段大小相符,可用于遗传转化实验。
将含阳性重组质粒的农杆菌接种于含抗生素的LB液体培养基中,制备转化液。采用花序侵染法侵染拟南芥突变体ttg1,获得了T0代转基因植株种子。将T0代种子进行抗性筛选,直到获得转基因纯合体株系。PCR检测验证了OsTTG1在转基因植株中的表达(图1:A),获得OsTTG1-OE拟南芥。利用RT-PCR检测OsTTG1在野生型、突变体以及过表达植株中的表达量,发现OsTTG1在过表达植株中稳定高表达(图1:B)。
2.2 拟南芥种子及植株表型分析
随机选取拟南芥3种基因型的成熟种子,在体式显微镜下观察种子形态。发现Col-0和OsTTG1-OE的种皮呈棕褐色,OsTTG1-OE的种皮颜色比野生型稍浅。突变体ttg1的种皮为浅黄色。取不同基因型的拟南芥种子各10粒,在体视显微镜下测量种子长度。结果显示,突变体ttg1的种子平均长度为0.46 mm,野生型为0.52 mm,OsTTG1-OE为0.53 mm,说明过表达OsTTG1后,拟南芥的种子大小受到影响(图2:A、B)。
选取苗龄10 d的拟南芥植株第1片真叶,在体视显微镜下观察表皮毛的密度,统计不同基因型拟南芥叶片上的表皮毛。结果显示,野生型拟南芥第1片真叶上表皮毛在35根左右,OsTTG1-OE在32根左右,突变体ttg1则几乎没有表皮毛(图2:C)。野生型和OsTTG1-OE植株的叶片、茎秆及花序上均有表皮毛,突变体ttg1则没有。此外,突变体ttg1几乎没有花青素的积累,过表达OsTTG1拟南芥株系则在叶片、茎等部位均有花青素的积累(图2:D)。
对不同基因型拟南芥的开花期进行观察,发现突变体ttg1的开花时间比Col-0和OsTTG1转基因株系提前3~5 d。Col-0和OsTTG1-OE的开花时间没有明显差异(图3),说明OsTTG1也能够影响拟南芥的开花时间。
2.3 RNA-seq分析结果
2.3.1 转录组测序分析
对拟南芥突变体ttg1和OsTTG1-OE株系共24个样品进行RNA-seq分析,共获得167.02 Gb 的Clean data,各样品的 Clean data均在6.01 Gb以上,Q30在93.51%以上,GC值均在46.01%以上。将各样品的Clean reads与参考基因组进行序列比对,发现匹配率在94.44%~96.47%。
2.3.2 差异表达基因统计分析
经统计,3个时期共得到
2424 个DEGs 。在莲座期突变体ttg1和OsTTG1-OE中筛选到1 913个DEGs,其中上调基因1 225个,包括可能与花青素积累有关的基因(MKK9、CYP75B1、GSTF12、DFRA、LDOX)、可能与表皮毛形成有关的基因(ETC2、SPL4、SPL3A),以及可能与调控开花有关的基因(FLC、HDA5)等;下调基因688个,包括可能与表皮毛形成有关的基因(SPL15、TCL1、ETC3)、可能与调控开花有关的基因(FT、AGL19、LHY)等。在抽薹期筛选到的DEGs为1 300个,其中上调基因752个,包括可能与花青素积累有关的基因(CYP75B1、TT8、GSTF12、F3H、CHS、DFRA、LDOX)、可能与表皮毛形成相关的基因(ETC2、SPL3A、GIS)等;下调基因548个,其中包括可能与表皮毛形成有关的基因ETC3。在结荚期筛选到差异表达基因1 008个,其中上调基因604个,包括可能与花青素积累有关的基因DFRA、LDOX,可能与表皮毛形成相关的基因ETC2、SPL3A;下调基因404个,包括可能与表皮毛形成相关的基因ETC3(图4:A)。对获得的DEGs进行分析,发现3个时期共有的DEGs为668个(图4:B)。对3个时期的DEGs进行层次聚类分析,发现多数在莲座期下调的基因,在抽薹期也下调,但在结荚期则上调;而在莲座期上调的基因,在其他两个时期则大多为下调(图4:C)。![]() 图 4 突变体ttg1与OsTTG1-OE株系3个时期的差异表达基因A:上调基因与下调基因统计;B:DEGs的韦恩图;C:DEGs的表达水平聚类热图。Stage1:莲座期;Stage2:抽薹期;Stage3:结荚期,下同。Figure 4. DEGs across three periods in Arabidopsis thaliana mutant ttg1 and OsTTG1-OEA: Statistical analysis of up-regulated and down-regulated genes; B: Venn diagram of DEGs; C: Heatmap of DEG expression levels. Stage 1: Rosette stage; Stage 2: Bolting stage; Stage 3: Bodding stage, same below.
图 4 突变体ttg1与OsTTG1-OE株系3个时期的差异表达基因A:上调基因与下调基因统计;B:DEGs的韦恩图;C:DEGs的表达水平聚类热图。Stage1:莲座期;Stage2:抽薹期;Stage3:结荚期,下同。Figure 4. DEGs across three periods in Arabidopsis thaliana mutant ttg1 and OsTTG1-OEA: Statistical analysis of up-regulated and down-regulated genes; B: Venn diagram of DEGs; C: Heatmap of DEG expression levels. Stage 1: Rosette stage; Stage 2: Bolting stage; Stage 3: Bodding stage, same below.2.3.3 差异表达基因GO(Gene ontology, GO)富集分析
对所有DEGs进行基因本体富集分析,发现在分子功能(Molecular function,MF)相关条目中,催化活性(GO: 0003824)、转录调节剂活性(GO: 0140110)、复合蛋白(GO: 0005488)等条目有较多的DEGs富集。在细胞组分(Cellular component,CC)相关条目中,膜部分(GO: 0044425)、细胞部分(GO: 0044464)、细胞器(GO: 0043226)等条目有较多的DEGs富集。在生物进程(Biological processes,BP)相关条目中,生物调节(GO: 0065007)、代谢过程(GO: 0008152)等条目有较多DEGs富集(图5)。在3个时期中,DEGs在催化活性、转录调节剂活性、代谢过程等相关条目均有较多富集。
3. 讨论
花青素是高等植物中的次生代谢产物,在抗逆、防御病虫害、育花、育种等方面发挥着重要作用[35]。花青素是在PAL、CHS、F3H、CHI、DFR、LDOX、UFGT等多种酶的催化下,在多种转录因子的调控下合成的[36]。在拟南芥中,当WD40的TTG1和bHLH的TT8、GL3或者EGL3与R2R3-MYB相互作用时,花青素的合成被激活。TTG1参与形成的MBW复合物能够直接与DFR、F3H、TT8、TTG2和ANS等基因的启动子结合,调控基因的表达。本研究中,水稻OsTTG1可以修复拟南芥突变体ttg1无花青素积累的表型缺陷。表明OsTTG1具有影响植株花青素积累的作用。RNA-seq分析发现了4个与花青素合成有关的酶,其中DFR是花青素合成途径中的关键酶,TTG1会影响DFR的功能,诱导DFR的表达,拟南芥突变体ttg1中DFR的催化受阻,花青素合成受到影响[37]。在拟南芥突变体ttg1中过表达OsTTG1后,发现3个时期的转基因植株中DFR的表达水平均比突变体ttg1高,说明OsTTG1可能也像TTG1一样能够诱导DFR的表达,影响花青素的积累。TT8属于bHLH转录因子,其能够与TT2(MYB)、TTG1(WD40)蛋白形成稳定的三元复合物,调控花青素合成途径中后期结构基因的表达,还能够通过正负反馈调节自身的表达,进而促进花青素的合成[38]。本研究中TT8基因的表达上调,说明OsTTG1或许也能与TT8及TT2形成复合物,调控花青素的合成。
表皮毛是拟南芥的一个重要性状,而突变体ttg1的表皮毛生长发育受阻,整个植株几乎没有表皮毛。本研究中,在拟南芥突变体ttg1中过表OsTTG1,可以修复突变体ttg1无表皮毛的表型缺陷,表明OsTTG1具有影响表皮毛形成的作用。RNA-seq分析发现,在抽薹期时,转基因植株中GIS的表达量显著高于突变体植株。GIS能够正调控拟南芥表皮毛的发育[22]。这说明OsTTG1可能通过调控GIS的表达来影响拟南芥表皮毛的形成。TCL1在拟南芥表皮毛形成过程中起着负调控的作用,AtTCL1过表达拟南芥植株的表皮毛形成被抑制[33]。我们发现,在莲座期时,转基因植株中GIS的表达量显著低于突变体植株,这说明OsTTG1可能对TCL1基因的表达量有影响,进而影响了莲座期表皮毛的形成。ETC2和TCL1一样,都是影响表皮毛形成的R3-MYB型负调控因子,但本研究发现,转基因植株中ETC2在3个时期的表达均显著上调。有研究指出,MBW对表皮毛的生长发育有正向调控作用,使R3-MYB型负调控因子TRY、CPC受到激活,使得MYB正向调控因子被取代,最后形成无活性的GY/CPC-GL3/EGL3-TTG1复合物[39]。我们猜测,在OsTTG1的影响下,MBW也能激活ETC2的表达,最终形成无活性的ETC2-GL3/EGL3-TTG1复合物,因此,ETC2的上调表达并没有抑制表皮毛的形成。
开花期是决定植物分布和季节适应性的重要农艺性状之一。本研究在拟南芥突变体ttg1中过表达OsTTG1基因,发现多个开花相关的基因只在莲座期表达量有差异,而在开花期的突变体ttg1和OsTTG1-OE植株中没有差异,说明OsTTG1可能会直接影响开花期,或者像TTG1一样可以调控下游生物合成相关的基因[17]。FLC是一个开花抑制基因,该基因表达上调会促使植株形成晚花表型[40],FLC的上调表达能够使拟南芥开花延迟。FT是拟南芥中的一种成花素,在花的形成过程中起着重要的作用。FT[29]和AGL19[41]基因的过表达均会导致拟南芥提前开花。本研究发现在莲座期时,转基因植株中FLC的表达量显著高于突变体植株,而FT和AGL19的表达量则显著下调。与开花相关的基因的表达量只在莲座期有差异,说明这些基因在植株抽薹前就能发挥功能。这3个基因的表达可能均受OsTTG1的影响,进而影响OsTTG1-OE植株的开花时间。有研究表明,TTG1过表达植株比野生型开花晚[5]。然而,本研究发现OsTTG1-OE植株的开花时间与野生型并没有明显差异。同时,我们发现在转基因植株中HDA5的表达量显著高于突变体植株。前人报道认为HDA5是一个正调控开花的基因,hda5突变体比野生型晚开花[42],与我们的研究结果刚好相反。因此,我们猜测OsTTG1可能会影响HDA5的表达,进而影响植株的开花时间。
在拟南芥突变体ttg1背景下,过表达OsTTG1株系的表型得到恢复,表明OsTTG1影响花青素生物合成、表皮毛形成及开花时间等性状。此外,TTG1对植株的根毛密度[43]、根长[44]、种皮黏液质[45]等也具有一定的影响。本研究中,RNA-seq分析发现的DEGs中也包括与氮素吸收利用、耐盐、抗病等相关的基因,因此,OsTTG1是否对上述性状也有影响还需进一步研究。
-
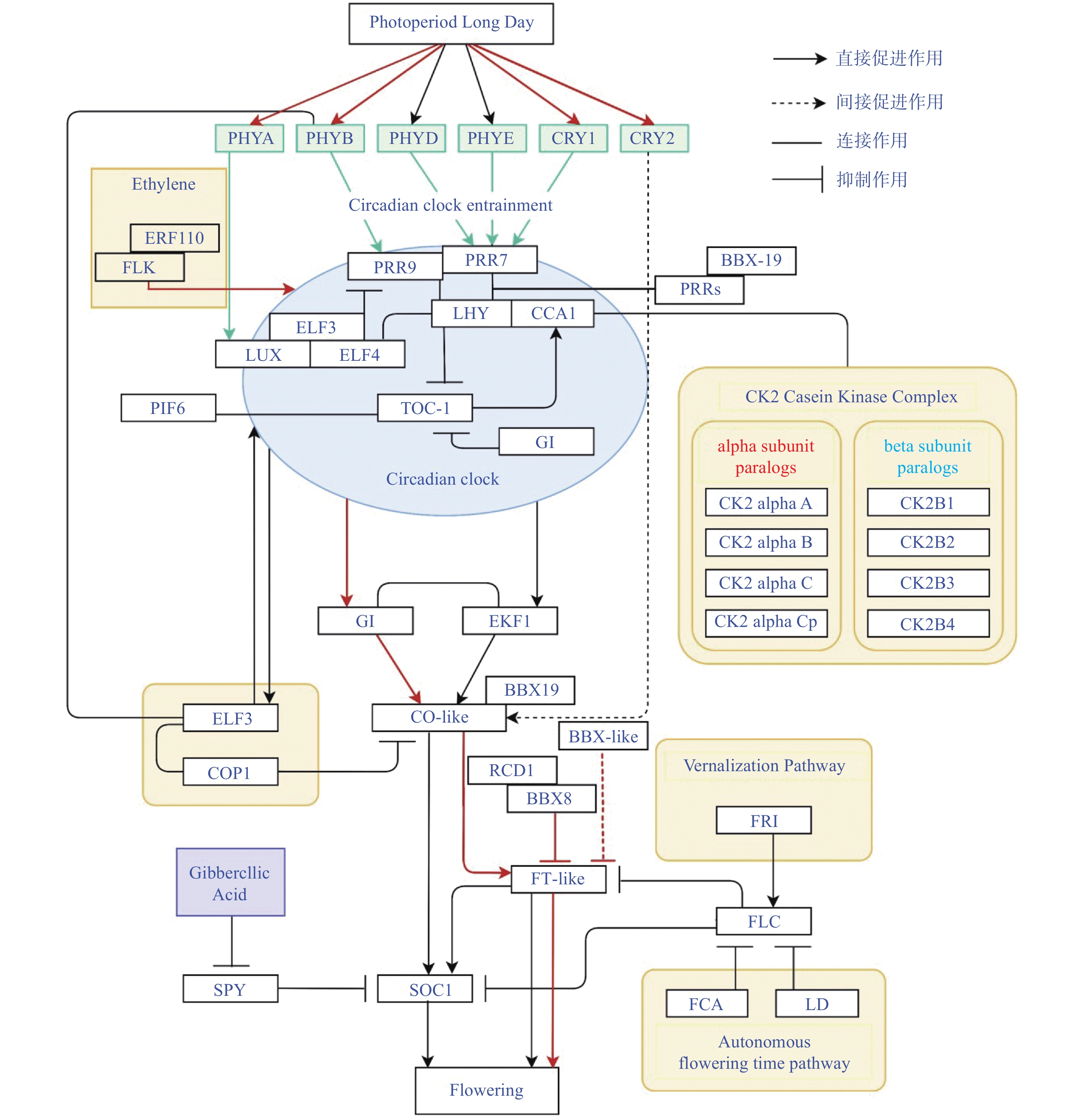
图 1 拟南芥中以光周期途径为主的开花途径和菊花中响应光周期的同源基因
红线代表在菊花中涉及的研究(参考网站:https://www.wikipathways.org/index.php/Pathway:WP622)。
Figure 1. Flowering pathway dominated by photoperiodic pathway in Arabidopsis thaliana and photoperiod-responsive homologous genes in Chrysanthemum
Red line represents research involving Chrysanthemum (Photoperiodic pathway available online: https://www.wikipathways.org/index.php/Pathway: WP2312).
-
[1] 舒黄英,郝园园,蔡庆泽,王振,朱国鹏,等. 模式植物拟南芥开花时间分子调控研究进展[J]. 植物科学学报,2017,35(4):603−608. doi: 10.11913/PSJ.2095-0837.2017.40603 Shu HY,Hao YY,Cai QZ,Wang Z,Zhu GP,et al. Recent research progress on the molecular regulation of flowering time in Arabidopsis thaliana[J]. Plant Science Journal,2017,35 (4):603−608. doi: 10.11913/PSJ.2095-0837.2017.40603
[2] 张秋玲,刘海鹏,高康,孔德元,戴思兰. 盆栽菊花反季节开花调控技术研究[J]. 黑龙江农业科学,2021(9):62−67. Zhang QL,Liu HP,Gao K,Kong DY,Dai SL. Research on anti-seasonal flowering control technology of potted chrysanthemum[J]. Heilongjiang Agricultural Sciences,2021 (9):62−67.
[3] 陈俊愉, 程绪珂. 富贵神仙品(中国花经节编)[M]//王明明. 大匠之门5. 南宁: 广西美术出版社, 2015: 1-100. [4] 谯德惠. 花卉产销实现平稳增长——2012年全国花卉统计数据分析[J]. 中国花卉园艺,2013(15):26−31. [5] 李小青. 我国花卉出口贸易的现状、问题及对策[J]. 中国市场,2022(29):75−78. doi: 10.13939/j.cnki.zgsc.2022.29.075 [6] 张引潮. 坚定信心锚定花卉业高质量发展——在2022全国花卉产销形势分析会上的讲话[J]. 中国花卉园艺,2022(4):10−15. doi: 10.3969/j.issn.1009-8496.2022.4.zghhyy202204003 [7] 尚嘉琪. 地被菊花期改良育种技术研究[D]. 晋中: 山西农业大学, 2017: 1-10. [8] 古晓红,李方舟,张海生,杨婷婷,王军. 大豆常规杂交育种和生物分子育种的优劣对比[J]. 种子科技,2020,38(17):29−30. doi: 10.3969/j.issn.1005-2690.2020.17.013 [9] 雒新艳,张俊丽,张二海. 盆栽小菊育种研究进展[J]. 山东林业科技,2021,51(1):81−86. doi: 10.3969/j.issn.1002-2724.2021.01.019 Luo XY,Zhang JL,Zhang EH. Research progress on potted chrysanthemum breeding[J]. Journal of Shandong Forestry Science and Technology,2021,51 (1):81−86. doi: 10.3969/j.issn.1002-2724.2021.01.019
[10] 赵小刚. 日中性小菊新品种选育及小菊开花期遗传分析[D]. 北京: 北京林业大学, 2019: 1-10. [11] 蒋志敏,王威,储成才. 植物氮高效利用研究进展和展望[J]. 生命科学,2018,30(10):1060−1071. doi: 10.13376/j.cbls/2018128 Jiang ZM,Wang W,Chu CC. Towards understanding of nitrogen use efficiency in plants[J]. Chinese Bulletin of Life Sciences,2018,30 (10):1060−1071. doi: 10.13376/j.cbls/2018128
[12] Gojon A. Nitrogen nutrition in plants:rapid progress and new challenges[J]. J Exp Bot,2017,68 (10):2457−2462. doi: 10.1093/jxb/erx171
[13] Srikanth A,Schmid M. Regulation of flowering time:all roads lead to Rome[J]. Cell Mol Life Sci,2011,68 (12):2013−2037. doi: 10.1007/s00018-011-0673-y
[14] Blümel M,Dally N,Jung C. Flowering time regulation in crops:what did we learn from Arabidopsis?[J]. Curr Opin Biotechnol,2015,32:121−129. doi: 10.1016/j.copbio.2014.11.023
[15] 孙昌辉,邓晓建,方军,储成才. 高等植物开花诱导研究进展[J]. 遗传,2007,29(10):1182−1190. doi: 10.3321/j.issn:0253-9772.2007.10.005 Sun CH,Deng XJ,Fang J,Chu CC. An overview of flowering transition in higher plants[J]. Hereditas (Beijing)
,2007,29 (10):1182−1190. doi: 10.3321/j.issn:0253-9772.2007.10.005 [16] 张艺能,周玉萍,陈琼华,黄小玲,田长恩. 拟南芥开花时间调控的分子基础[J]. 植物学报,2014,49(4):469−482. doi: 10.3724/SP.J.1259.2014.00469 Zhang YN,Zhou YP,Chen QH,Huang XL,Tian CE. Molecular basis of flowering time regulation in Arabidopsis[J]. Chinese Bulletin of Botany,2014,49 (4):469−482. doi: 10.3724/SP.J.1259.2014.00469
[17] Komeda Y. Genetic regulation of time to flower in Arabidopsis thaliana[J]. Annu Rev Plant Biol,2004,55:521−535. doi: 10.1146/annurev.arplant.55.031903.141644
[18] Wahl V,Ponnu J,Schlereth A,Arrivault S,Langenecker T,et al. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana[J]. Science,2013,339 (6120):704−707. doi: 10.1126/science.1230406
[19] Teotia S,Tang GL. To bloom or not to bloom:role of MicroRNAs in plant flowering[J]. Mol Plant,2015,8 (3):359−377. doi: 10.1016/j.molp.2014.12.018
[20] Achard P,Cheng H,de Grauwe L,Decat J,Schoutteten H,et al. Integration of plant responses to environmentally activated phytohormonal signals[J]. Science,2006,311 (5757):91−94. doi: 10.1126/science.1118642
[21] Balasubramanian S,Sureshkumar S,Lempe J,Weigel D. Potent induction of Arabidopsis thaliana flowering by elevated growth temperature[J]. PLoS Genetics,2006,2 (7):e106. doi: 10.1371/journal.pgen.0020106
[22] Martínez C,Pons E,Prats G,León J. Salicylic acid regulates flowering time and links defence responses and reproductive development[J]. Plant J,2004,37 (2):209−217. doi: 10.1046/j.1365-313X.2003.01954.x
[23] Vidal EA,Moyano TC,Canales J,Gutiérrez RA. Nitrogen control of developmental phase transitions in Arabidopsis thaliana[J]. J Exp Bot,2014,65 (19):5611−5618. doi: 10.1093/jxb/eru326
[24] Kitamoto N,Ueno S,Takenaka A,Tsumura Y,Washitani I,Ohsawa R. Effect of flowering phenology on pollen flow distance and the consequences for spatial genetic structure within a population of Primula sieboldii (Primulaceae)[J]. Am J Bot,2006,93 (2):226−233. doi: 10.3732/ajb.93.2.226
[25] Elzinga JA,Atlan A,Biere A,Gigord L,Weis AE,Bernasconi G. Time after time:flowering phenology and biotic interactions[J]. Trends Ecol Evol,2007,22 (8):432−439. doi: 10.1016/j.tree.2007.05.006
[26] Lemoine NP,Doublet D,Salminen JP,Burkepile DE,Parker JD. Responses of plant phenology,growth,defense,and reproduction to interactive effects of warming and insect herbivory[J]. Ecology,2017,98 (7):1817−1828. doi: 10.1002/ecy.1855
[27] Vermeulen PJ. On selection for flowering time plasticity in response to density[J]. New Phytol,2015,205 (1):429−439. doi: 10.1111/nph.12984
[28] 万亚楠. 菊花的花期调控方法初探[J]. 现代园艺,2013(20):50−51. doi: 10.3969/j.issn.1006-4958.2013.20.035 [29] 张树林, 戴思兰. 中国菊花全书[M]. 北京: 中国林业出版社, 2013: 1-100. [30] 陈洪国,马容明. GA3对菊花开花和花瓣某些生理生化指标的影响[J]. 安徽农业科学,2006,34(6):1050−1051. doi: 10.3969/j.issn.0517-6611.2006.06.005 Chen HG,Ma RM. Effects of GA3 on the flowering and some physiological indexes of chrysanthemum[J]. Journal of Anhui Agricultural Sciences,2006,34 (6):1050−1051. doi: 10.3969/j.issn.0517-6611.2006.06.005
[31] 刘敏,丁江南,王飞翔,于晓英. 叶面喷施赤霉素对瓜叶菊生长与开花的影响[J]. 天津农业科学,2010,16(6):36−38. doi: 10.3969/j.issn.1006-6500.2010.06.014 Liu M,Ding JN,Wang FX,Yu XY. Effects of gibberellins treatment on growth and flowering of Senecio × hybridus[J]. Tianjin Agricultural Sciences,2010,16 (6):36−38. doi: 10.3969/j.issn.1006-6500.2010.06.014
[32] 张秋玲,杨秀珍,戴思兰,张倩,罗虹,张伯晗. 不同氮磷钾水平对毛华菊生长发育的影响[J]. 山东农业大学学报(自然科学版),2020,51(4):611−616. doi: 10.3969/j.issn.1000-2324.2020.04.005 Zhang QL,Yang XZ,Dai SL,Zhang Q,Luo H,Zhang BH. Effect of different N,P,K proportions on the development of Chrysanthemum vestitum[J]. Journal of Shandong Agricultural University (Natural Science Edition)
,2020,51 (4):611−616. doi: 10.3969/j.issn.1000-2324.2020.04.005 [33] 张秋玲,杨秀珍,戴思兰,邱丹丹,董南希,李清清. 不同氮水平下毛华菊形态性状的差异分析[J]. 中国农业大学学报,2020,25(5):70−77. doi: 10.11841/j.issn.1007-4333.2020.05.07 Zhang QL,Yang XZ,Dai SL,Qiu DD,Dong NX,Li QQ. Difference analysis of morphological traits of Chrysanthemum vestitum under different nitrogen levels[J]. Journal of China Agricultural University,2020,25 (5):70−77. doi: 10.11841/j.issn.1007-4333.2020.05.07
[34] 马朝峰. 甘菊和毛华菊PHYA和PHYB同源基因表达分析及ClPHYB功能验证[D]. 北京: 北京林业大学, 2019: 1-10. [35] 王富刚,张静,张雄. 光敏色素与植物的光形态建成[J]. 基因组学与应用生物学,2017,36(8):3167−3171. doi: 10.13417/j.gab.036.003167 Wang FG,Zhang J,Zhang X. Phytochromes and plant photomorphogenesis[J]. Genomics and Applied Biology,2017,36 (8):3167−3171. doi: 10.13417/j.gab.036.003167
[36] 王君杰,田翔,秦慧彬,王海岗,曹晓宁,等. 光周期对糜子生长发育及叶片内源激素的调控效应[J]. 中国农业科学,2021,54(2):286−295. doi: 10.3864/j.issn.0578-1752.2021.02.005 Wang JJ,Tian X,Qin HB,Wang HG,Cao XN,et al. Regulation effects of photoperiod on growth and leaf endogenous hormones in broomcorn millet[J]. Scientia Agricultura Sinica,2021,54 (2):286−295. doi: 10.3864/j.issn.0578-1752.2021.02.005
[37] 贺玉利. 菊花矮化及提前开花栽培技术[J]. 北方园艺,2003(3):80. doi: 10.3969/j.issn.1001-0009.2003.03.055 [38] 姜贝贝,房伟民,陈发棣,赵宏波,顾俊杰. 植株营养生长天数对切花菊花芽分化与品质的影响[J]. 中国农业科学,2008,41(6):1755−1760. doi: 10.3864/j.issn.0578-1752.2008.06.024 Jiang BB,Fang WM,Chen FD,Zhao HB,Gu JJ. Effect of vegetative growth days on flower bud differentiation and quality of cut chrysanthemum[J]. Scientia Agricultura Sinica,2008,41 (6):1755−1760. doi: 10.3864/j.issn.0578-1752.2008.06.024
[39] 陆思宇,杨再强,杨立,张源达,郑涵. 不同光周期对菊花生长发育及内源激素的影响[J]. 华北农学报,2021,36(6):106−115. doi: 10.7668/hbnxb.20192386 Lu SY,Yang ZQ,Yang L,Zhang YD,Zheng H. Effects of different photoperiods on the growth and development process and endogenous hormones of chrysanthemum[J]. Acta Agriculturae Boreali-Sinica,2021,36 (6):106−115. doi: 10.7668/hbnxb.20192386
[40] 陆思宇. 光周期对‘红面’菊花生长发育的影响机理[D]. 南京: 南京信息工程大学, 2022: 1-10. [41] Hsu PY,Harmer SL. Wheels within wheels:the plant circadian system[J]. Trends Plant Sci,2014,19 (4):240−249. doi: 10.1016/j.tplants.2013.11.007
[42] Yu JW,Rubio V,Lee NY,Bai SL,Lee SY,et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability[J]. Molecular Cell,2008,32 (5):617−630. doi: 10.1016/j.molcel.2008.09.026
[43] Song YH,Shim JS,Kinmonth-Schultz HA,Imaizumi T. Photoperiodic flowering:time measurement mechanisms in leaves[J]. Annu Rev Plant Biol,2015,66:441−464. doi: 10.1146/annurev-arplant-043014-115555
[44] Shim JS,Kubota A,Imaizumi T. Circadian clock and photoperiodic flowering in Arabidopsis:CONSTANS is a hub for signal integration[J]. Plant Physiol,2017,173 (1):5−15. doi: 10.1104/pp.16.01327
[45] Jing YJ,Guo Q,Lin RC. The chromatin-remodeling factor PICKLE antagonizes polycomb repression of FT to promote flowering[J]. Plant Physiol,2019,181 (2):656−668. doi: 10.1104/pp.19.00596
[46] Jing YJ,Guo Q,Zha P,Lin RC. The chromatin‐remodelling factor PICKLE interacts with CONSTANS to promote flowering in Arabidopsis[J]. Plant Cell Environ,2019,42 (8):2495−2507. doi: 10.1111/pce.13557
[47] Kadman-Zahavi A,Yahel H. Phytochrome effects in night-break illuminations on flowering of Chrysanthemum[J]. Physiol Plant,1971,25 (1):90−93. doi: 10.1111/j.1399-3054.1971.tb01094.x
[48] Jeong SW,Park S,Jin JS,Seo ON,Kim GS,et al. Influences of four different light-emitting diode lights on flowering and polyphenol variations in the leaves of chrysanthemum (Chrysanthemum morifolium)[J]. J Agric Food Chem,2012,60 (39):9793−9800. doi: 10.1021/jf302272x
[49] Nissim-Levi A,Kitron M,Nishri Y,Ovadia R,Forer I,et al. Effects of blue and red LED lights on growth and flowering of Chrysanthemum morifolium[J]. Sci Hortic,2019,254:77−83. doi: 10.1016/j.scienta.2019.04.080
[50] Yang LW,Wen XH,Fu JX,Dai SL. ClCRY2 facilitates floral transition in Chrysanthemum lavandulifolium by affecting the transcription of circadian clock-related genes under short-day photoperiods[J]. Hortic Res,2018,5:58. doi: 10.1038/s41438-018-0063-9
[51] Yang LW,Fu JX,Qi S,Hong Y,Huang H,Dai SL. Molecular cloning and function analysis of ClCRY1a and ClCRY1b,two genes in Chrysanthemum lavandulifolium that play vital roles in promoting floral transition[J]. Gene,2017,617:32−43. doi: 10.1016/j.gene.2017.02.020
[52] Wang SJ,Zhang CL,Zhao J,Li RH,Lv JH. Expression analysis of four pseudo-response regulator (PRR) genes in Chrysanthemum morifolium under different photoperiods[J]. PeerJ,2019,7:e6420. doi: 10.7717/peerj.6420
[53] 陈丹丹,邹庆军,郭巧生,汪涛. 短日照处理对野菊CO基因表达量的影响[J]. 中国中药杂志,2019,44(4):648−653. doi: 10.19540/j.cnki.cjcmm.2019.0014 Chen DD,Zou QJ,Guo QS,Wang T. Effect of short-day treatment on expression of CO gene in Chrysanthemum indicum[J]. China Journal of Chinese Materia Medica,2019,44 (4):648−653. doi: 10.19540/j.cnki.cjcmm.2019.0014
[54] Oda A,Narumi T,Li TP,Kando T,Higuchi Y,et al. CsFTL3,a chrysanthemum FLOWERING LOCUS T-like gene,is a key regulator of photoperiodic flowering in chrysanthemums[J]. J Exp Bot,2012,63 (3):1461−1477. doi: 10.1093/jxb/err387
[55] Higuchi Y,Narumi T,Oda A,Nakano Y,Sumitomo K,et al. The gated induction system of a systemic floral inhibitor,antiflorigen,determines obligate short-day flowering in chrysanthemums[J]. Proc Natl Acad Sci USA,2013,110 (42):17137−17142. doi: 10.1073/pnas.1307617110
[56] Sun J,Wang H,Ren LP,Chen SM,Chen FD,et al. CmFTL2 is involved in the photoperiod- and sucrose-mediated control of flowering time in chrysanthemum[J]. Hortic Res,2017,4:17001. doi: 10.1038/hortres.2017.1
[57] Zuo L,Wang T,Guo Q,Yang F,Zou Q,et al. Conserved CO-FT module regulating flowering time in Chrysanthemum indicum L.[J]. Russ J Plant Physiol,2021,68 (6):1018−1028. doi: 10.1134/S102144372106025X
[58] Oda A,Higuchi Y,Hisamatsu T. Photoperiod-insensitive floral transition in chrysanthemum induced by constitutive expression of chimeric repressor CsLHY-SRDX[J]. Plant Sci,2017,259:86−93. doi: 10.1016/j.plantsci.2017.03.007
[59] Oda A,Higuchi Y,Hisamatsu T. Constitutive expression of CsGI alters critical night length for flowering by changing the photo-sensitive phase of anti-florigen induction in chrysanthemum[J]. Plant Sci,2020,293:110417. doi: 10.1016/j.plantsci.2020.110417
[60] 赵航, 梁丽, 张淑欣. 温度调控植物开花的研究进展[J/OL]. 分子植物育种, 2022. https: //kns.cnki.net/kcms/detail/46.1068.S.20220420.1718.020.html. Zhao H, Liang L, Zhang SX. Research progress on temperature-regulated of plant flowering[J]. Molecular Plant Breeding, 2022. https: //kns.cnki.net/kcms/detail/46.1068.S.20220420.1718.020.html.
[61] Laurie DA. Comparative genetics of flowering time[J]. Plant Mol Biol,1997,35 (1-2):167−177.
[62] Trevaskis B,Hemming MN,Dennis ES,Peacock WJ. The molecular basis of vernalization-induced flowering in cereals[J]. Trends Plant Sci,2007,12 (8):352−357. doi: 10.1016/j.tplants.2007.06.010
[63] Bouché F,Woods DP,Amasino RM. Winter memory throughout the plant kingdom:different paths to flowering[J]. Plant Physiol,2017,173 (1):27−35. doi: 10.1104/pp.16.01322
[64] Kim DH,Sung S. Vernalization-triggered intragenic chromatin loop formation by long noncoding RNAs[J]. Dev Cell,2017,40 (3):302−312.e4. doi: 10.1016/j.devcel.2016.12.021
[65] Xu SJ,Xiao J,Yin F,Guo XY,Xing LJ,et al. The protein modifications of O-GlcNAcylation and phosphorylation mediate vernalization response for flowering in winter wheat[J]. Plant Physiol,2019,180 (3):1436−1449. doi: 10.1104/pp.19.00081
[66] Lutz U,Nussbaumer T,Spannagl M,Diener J,Mayer KF,Schwechheimer C. Natural haplotypes of FLM non-coding sequences fine-tune flowering time in ambient spring temperatures in Arabidopsis[J]. eLife,2017,6:e22114. doi: 10.7554/eLife.22114
[67] Kumar SV,Lucyshyn D,Jaeger KE,Alós E,Alvey E,et al. Transcription factor PIF4 controls the thermosensory activation of flowering[J]. Nature,2012,484 (7393):242−245. doi: 10.1038/nature10928
[68] Song YH,Ito S,Imaizumi T. Flowering time regulation:photoperiod- and temperature-sensing in leaves[J]. Trends Plant Sci,2013,18 (10):575−583. doi: 10.1016/j.tplants.2013.05.003
[69] Jin SY,Ahn JH. Regulation of flowering time by ambient temperature:repressing the repressors and activating the activators[J]. New Phytol,2021,230 (3):938−942. doi: 10.1111/nph.17217
[70] Posé D,Verhage L,Ott F,Yant L,Mathieu J,et al. Temperature-dependent regulation of flowering by antagonistic FLM variants[J]. Nature,2013,503 (7476):414−417. doi: 10.1038/nature12633
[71] Kim JJ,Lee JH,Kim W,Jung HS,Huijser P,Ahn JH. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis[J]. Plant Physiol,2012,159 (1):461−478. doi: 10.1104/pp.111.192369
[72] Jung JH,Seo PJ,Ahn JH,Park CM. Arabidopsis RNA-binding protein FCA regulates MicroRNA172 processing in thermosensory flowering[J]. J Biol Chem,2012,287 (19):16007−16016. doi: 10.1074/jbc.M111.337485
[73] Kumar SV,Wigge PA. H2A. Z-containing nucleosomes mediate the thermosensory response in Arabidopsis[J]. Cell,2010,140 (1):136−147. doi: 10.1016/j.cell.2009.11.006
[74] Zheng SZ,Hu HM,Ren HM,Yang ZL,Qiu Q,et al. The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor[J]. Nat Commun,2019,10 (1):1303. doi: 10.1038/s41467-019-09310-x
[75] Huang H,Nusinow DA. Into the evening:complex interactions in the Arabidopsis circadian clock[J]. Trends Genet,2016,32 (10):674−686. doi: 10.1016/j.tig.2016.08.002
[76] Ezer D,Jung JH,Lan H,Biswas S,Gregoire L,et al. The evening complex coordinates environmental and endogenous signals in Arabidopsis[J]. Nat Plants,2017,3 (7):17087. doi: 10.1038/nplants.2017.87
[77] Zhao H,Xu D,Tian T,Kong FY,Lin K,et al. Molecular and functional dissection of EARLY-FLOWERING 3 (ELF3) and ELF4 in Arabidopsis[J]. Plant Sci,2021,303:110786. doi: 10.1016/j.plantsci.2020.110786
[78] Cho AR,Kim YJ. Night temperature determines flowering time and quality of Chrysanthemum morifolium during a high day temperature[J]. J Hortic Sci Biotechnol,2021,96 (2):239−248. doi: 10.1080/14620316.2020.1834460
[79] Cockshull KE,Kofranek AM. High night temperatures delay flowering,produce abnormal flowers and retard stem growth of cut-flower chrysanthemums[J]. Sci Hortic,1994,56 (3):217−234. doi: 10.1016/0304-4238(94)90004-3
[80] Nakano Y,Higuchi Y,Sumitomo K,Oda A,Hisamatsu T,Naro N. Delay of flowering by high temperature in chrysanthemum:heat-sensitive time-of-day and heat effects on CsFTL3 and CsAFT gene expression[J]. J Hortic Sci Biotechnol,2015,90 (2):143−149. doi: 10.1080/14620316.2015.11513165
[81] Nakano Y,Takase T,Sumitomo K,Suzuki S,Tsuda-Kawamura K,Hisamatsu T. Delay of flowering at high temperature in chrysanthemum:duration of darkness and transitions in lighting determine daily peak heat sensitivity[J]. Hortic J,2020,89 (5):602−608. doi: 10.2503/hortj.UTD-192
[82] Luo C,Liu H,Ren JN,Chen DL,Cheng X,et al. Cold-inducible expression of an Arabidopsis thaliana AP2 transcription factor gene,AtCRAP2,promotes flowering under unsuitable low-temperatures in chrysanthemum[J]. Plant Physiol Biochem,2020,146:220−230. doi: 10.1016/j.plaphy.2019.11.022
[83] Lyu J,Aiwaili P,Gu ZY,Xu YJ,Zhang YH,et al. Chrysanthemum MAF2 regulates flowering by repressing gibberellin biosynthesis in response to low temperature[J]. Plant J,2022,112 (5):1159−1175. doi: 10.1111/tpj.16002
[84] Sumitomo K,Nakano Y,Hisamatsu T,Oda A,Narumi-Kawasaki T,et al. Delayed flowering due to ‘cold memory’ is regulated by suppression of FLOWERING LOCUS T-like 3 gene in chrysanthemums[J]. J Hortic Sci Biotechnol,2023,98 (3):334−341. doi: 10.1080/14620316.2022.2136112
[85] Zhang XY,Zhang P,Wang G,Bao ZL,Ma FF. Chrysanthemum lavandulifolium homolog ClMAD1 modulates the floral transition during temperature shift[J]. Environ Exp Bot,2022,194:104720. doi: 10.1016/j.envexpbot.2021.104720
[86] Sumitomo K,Li TP,Hisamatsu T. Gibberellin promotes flowering of chrysanthemum by upregulating CmFL,a chrysanthemum FLORICAULA/LEAFY homologous gene[J]. Plant Sci,2009,176 (5):643−649. doi: 10.1016/j.plantsci.2009.02.003
[87] Wilson RN,Heckman JW,Somerville CR. Gibberellin is required for flowering in Arabidopsis thaliana under short days[J]. Plant Physiol,1992,100 (1):403−408. doi: 10.1104/pp.100.1.403
[88] Murase K,Hirano Y,Sun TP,Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1[J]. Nature,2008,456 (7221):459−463. doi: 10.1038/nature07519
[89] Yan JD,Li XM,Zeng BJ,Zhong M,Yang JX,et al. FKF1 F‐box protein promotes flowering in part by negatively regulating DELLA protein stability under long‐day photoperiod in Arabidopsis[J]. J Integr Plant Biol,2020,62 (11):1717−1740. doi: 10.1111/jipb.12971
[90] Achard P,Herr A,Baulcombe DC,Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA[J]. Development,2004,131 (14):3357−3365. doi: 10.1242/dev.01206
[91] Allen RS,Li JY,Stahle MI,Dubroué A,Gubler F,Millar AA. Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family[J]. Proc Natl Acad Sci USA,2007,104 (41):16371−16376. doi: 10.1073/pnas.0707653104
[92] Pharis RP. Flowering of Chrysanthemum under non-inductive long days by gibberellins and N6-benzyladenine[J]. Planta,1972,105 (3):205−212. doi: 10.1007/BF00385392
[93] Dong B,Deng Y,Wang HB,Gao R,Stephen GU,et al. Gibberellic acid signaling is required to induce flowering of chrysanthemums grown under both short and long days[J]. Int J Mol Sci,2017,18 (6):1259. doi: 10.3390/ijms18061259
[94] Yang YJ,Ma C,Xu YJ,Wei Q,Imtiaz M,et al. A zinc finger protein regulates flowering time and abiotic stress tolerance in chrysanthemum by modulating gibberellin biosynthesis[J]. Plant Cell,2014,26 (5):2038−2054. doi: 10.1105/tpc.114.124867
[95] Zhu L,Guan YX,Liu YN,Zhang ZH,Jaffar MA,et al. Regulation of flowering time in chrysanthemum by the R2R3 MYB transcription factor CmMYB2 is associated with changes in gibberellin metabolism[J]. Hortic Res,2020,7 (1):96. doi: 10.1038/s41438-020-0317-1
[96] Wu G,Park MY,Conway SR,Wang JW,Weigel D,Poethig RS. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis[J]. Cell,2009,138 (4):750−759. doi: 10.1016/j.cell.2009.06.031
[97] Wang JW,Czech B,Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana[J]. Cell,2009,138 (4):738−749. doi: 10.1016/j.cell.2009.06.014
[98] Fornara F,Coupland G. Plant phase transitions make a SPLash[J]. Cell,2009,138 (4):625−627. doi: 10.1016/j.cell.2009.08.011
[99] Yang HC,Han ZF,Cao Y,Fan D,Li H,et al. A companion cell–dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression[J]. PLoS Genet,2012,8 (4):e1002664. doi: 10.1371/journal.pgen.1002664
[100] Song AP,Gao TW,Wu D,Xin JJ,Chen SM,et al. Transcriptome-wide identification and expression analysis of chrysanthemum SBP-like transcription factors[J]. Plant Physiol Biochem,2016,102:10−16. doi: 10.1016/j.plaphy.2016.02.009
[101] 朱文静. 菊花转录因子CmSPL4. 1/5. 1/6/13的克隆与功能鉴定[D]. 南京: 南京农业大学, 2020: 1-10. [102] 魏倩. 菊花核因子NF-YB调节开花时间和干旱胁迫耐性的机理分析[D]. 北京: 中国农业大学, 2015: 1-10. [103] Wei Q,Ma C,Xu YJ,Wang TL,Chen YY,et al. Control of chrysanthemum flowering through integration with an aging pathway[J]. Nat Commun,2017,8 (1):829. doi: 10.1038/s41467-017-00812-0
[104] 马超. 菊花成花调控机制: 第三届全国植物开花·衰老与采后生物学大会论文摘要集[C]. 杭州: 中国植物生理与植物分子生物学学会, 2019. [105] Jiang JF, Zhang ZX, Hu Q, Zhu YQ, Gao Z, et al. The flowering repressor SVP recruits the TOPLESS co-repressor to control flowering in chrysanthemum and Arabidopsis[J/OL]. BioRxiv, 2021. doi: 10.1101/2021.11.23.469726.
[106] Wang CQ,Guthrie C,Sarmast MK,Dehesh K. BBX19 interacts with CONSTANS to repress FLOWERING LOCUS T transcription,defining a flowering time checkpoint in Arabidopsis[J]. Plant Cell,2014,26 (9):3589−3602. doi: 10.1105/tpc.114.130252
[107] Yuan L,Yu YJ,Liu MM,Song Y,Li HM,et al. BBX19 fine-tunes the circadian rhythm by interacting with PSEUDO-RESPONSE REGULATOR proteins to facilitate their repressive effect on morning-phased clock genes[J]. Plant Cell,2021,33 (8):2602−2617. doi: 10.1093/plcell/koab133
[108] Zhang T. Tick-tock:BBX19 functions as a novel regulator of the circadian clock[J]. Plant Cell,2021,33 (8):2511−2512. doi: 10.1093/plcell/koab142
[109] Wang LJ,Sun J,Ren LP,Zhou M,Han XY,et al. CmBBX8 accelerates flowering by targeting CmFTL1 directly in summer chrysanthemum[J]. Plant Biotechnol J,2020,18 (7):1562−1572. doi: 10.1111/pbi.13322
[110] Wang LJ,Cheng H,Wang Q,Si CN,Yang YM,et al. CmRCD1 represses flowering by directly interacting with CmBBX8 in summer chrysanthemum[J]. Hortic Res,2021,8:79. doi: 10.1038/s41438-021-00516-z
[111] Chen H,Huang F,Liu YN,Cheng PL,Guan ZY,et al. Constitutive expression of chrysanthemum CmBBX29 delays flowering time in transgenic Arabidopsis[J]. Can J Plant Sci,2020,100 (1):86−94. doi: 10.1139/cjps-2018-0154
[112] Ping Q,Cheng PL,Huang F,Ren LP,Cheng H,et al. The heterologous expression in Arabidopsis thaliana of a chrysanthemum gene encoding the BBX family transcription factor CmBBX13 delays flowering[J]. Plant Physiol Biochem,2019,144:480−487. doi: 10.1016/j.plaphy.2019.10.019
[113] Morita S,Murakoshi Y,Hojo A,Chisaka K,Harada T,Satoh S. Early flowering and increased expression of a FLOWERING LOCUS T-like gene in chrysanthemum transformed with a mutated ethylene receptor gene mDG-ERS1(etr1-4)[J]. J Plant Biol,2012,55 (5):398−405. doi: 10.1007/s12374-012-0109-8
[114] Huang YY,Xing XJ,Tang Y,Jin JY,Ding L,et al. An ethylene‐responsive transcription factor and a flowering locus KH domain homologue jointly modulate photoperiodic flowering in chrysanthemum[J]. Plant Cell Environ,2022,45 (5):1442−1456. doi: 10.1111/pce.14261
[115] Gomi K. Jasmonic acid:an essential plant hormone[J]. Int J Mol Sci,2020,21 (4):1261. doi: 10.3390/ijms21041261
[116] Guan YX,Ding L,Jiang JF,Shentu YY,Zhao WQ,et al. Overexpression of the CmJAZ1-like gene delays flowering in Chrysanthemum morifolium[J]. Hortic Res,2021,8:87. doi: 10.1038/s41438-021-00525-y
[117] Yuan S,Zhang ZW,Zheng C,Zhao ZY,Wang Y,et al. Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering[J]. Proc Natl Acad Sci USA,2016,113 (27):7661−7666. doi: 10.1073/pnas.1602004113
[118] Lin YL,Tsay YF. Influence of differing nitrate and nitrogen availability on flowering control in Arabidopsis[J]. J Exp Bot,2017,68 (10):2603−2609. doi: 10.1093/jxb/erx053
[119] Sanagi M,Aoyama S,Kubo A,Lu Y,Sato Y,et al. Low nitrogen conditions accelerate flowering by modulating the phosphorylation state of FLOWERING BHLH 4 in Arabidopsis[J]. Proc Natl Acad Sci USA,2021,118 (19):e2022942118. doi: 10.1073/pnas.2022942118
[120] Zhang SN,Zhang YY,Li KN,Yan M,Zhang JF,et al. Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice[J]. Curr Biol,2021,31 (4):671−683.e5. doi: 10.1016/j.cub.2020.10.095




 下载:
下载: