Construction of two-hybrid library of yeast and screening of NnWRKY40 interacting proteins in Nelumbo nucifera Gaertn. ‘Baihuajian’
-
摘要:
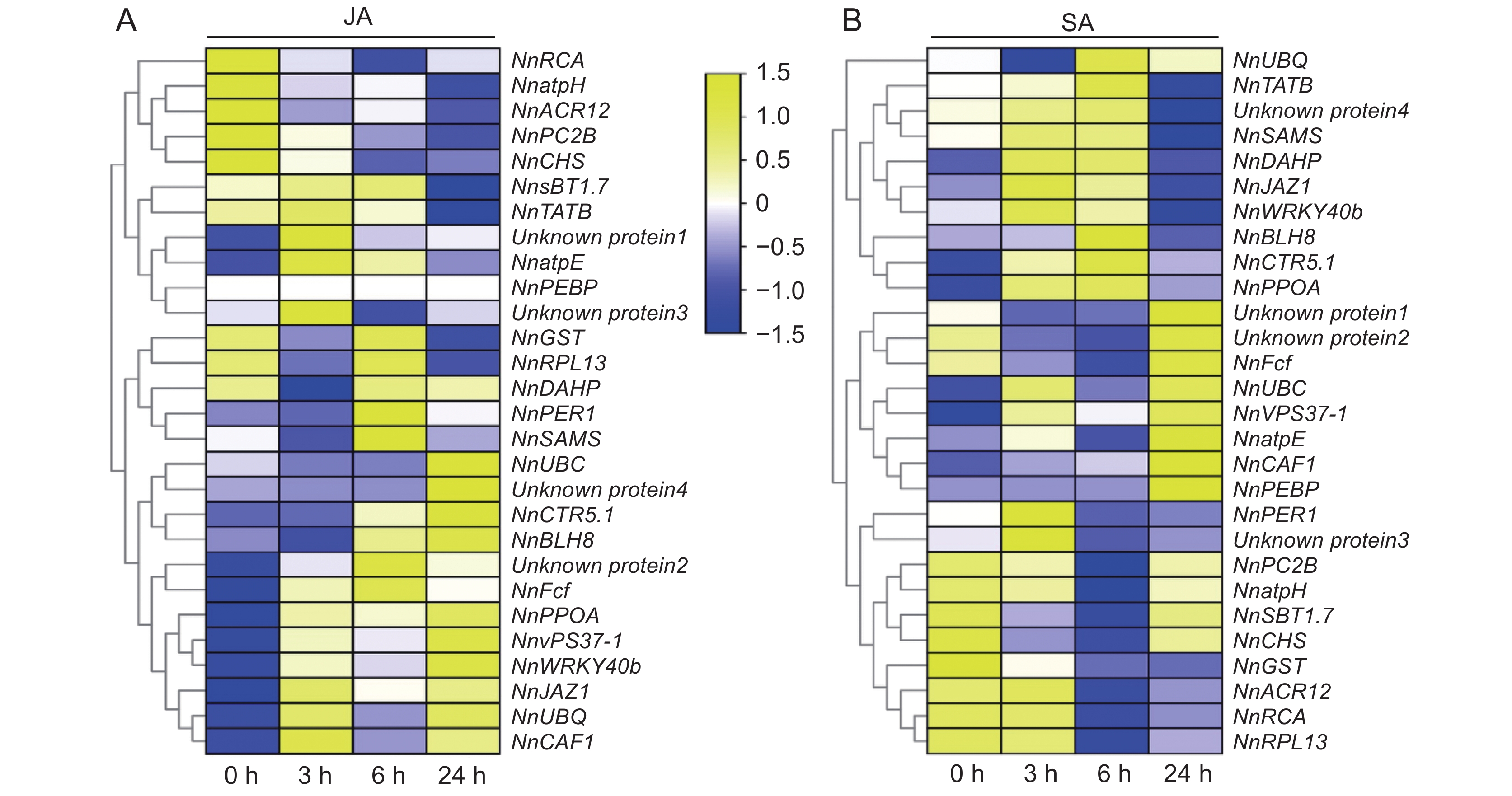
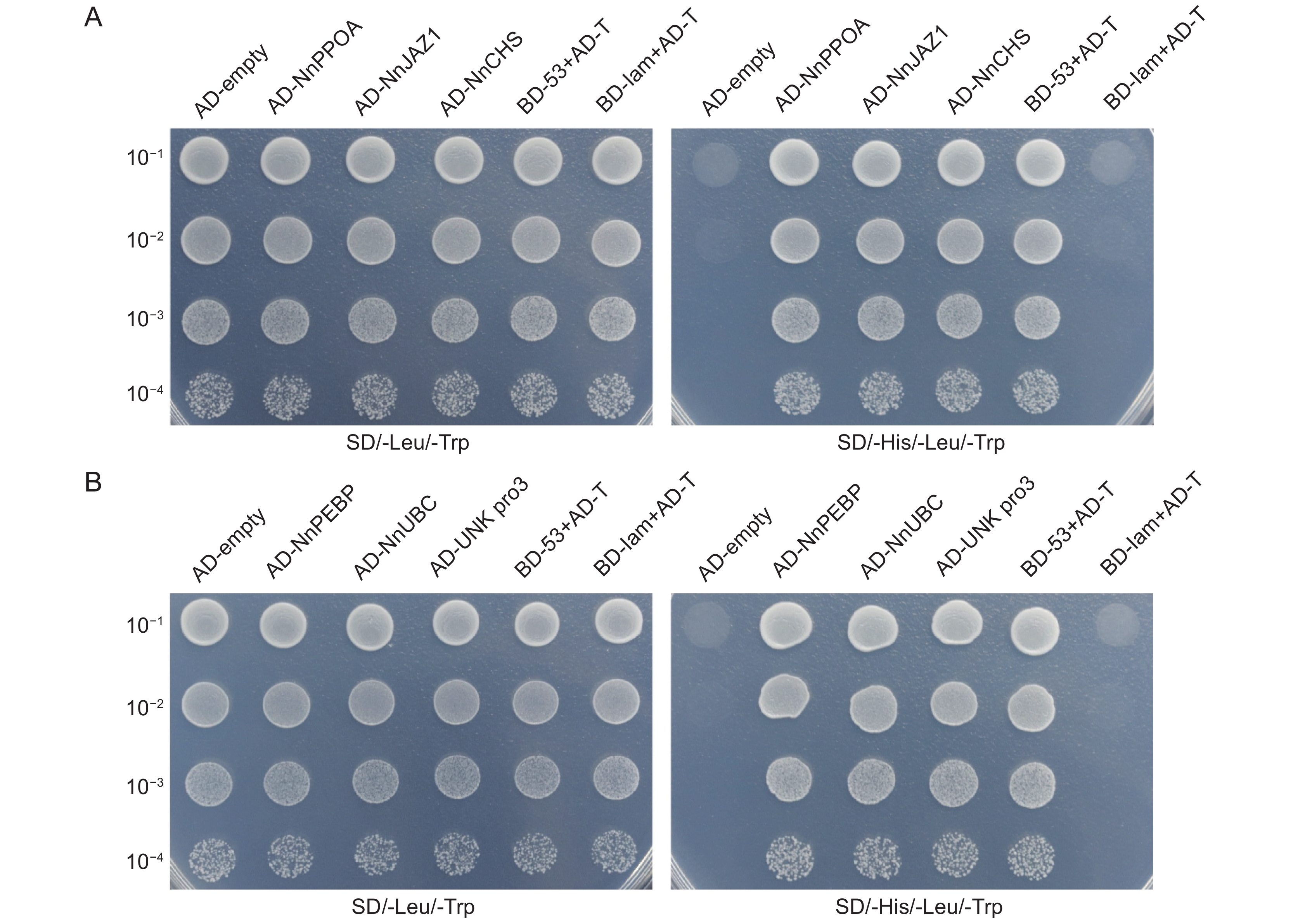
通过构建莲(Nelumbo nucifera Gaertn.)品种 ‘白花建’不同组织的混合cDNA文库,筛选与莲转录因子NnWRKY40互作的蛋白,探究NnWRKY40参与调控次级代谢物生物碱合成的可能机制。结果显示,混合cDNA文库的库容为1.2 × 107 CFU,重组率为100%,插入片段平均长度大于1000 bp。NnWRKY40包含两个同源基因NnWRKY40a和NnWRKY40b,利用NnWRKY40b构建诱饵载体pGBKT7-NnWRKY40b,通过共转化方法,从文库中筛选到27个与NnWRKY40b 互作的蛋白。这些互作蛋白可分为生长发育及抗逆、激素调控和次级代谢、未知蛋白3类。选取6个代表性互作蛋白(NnUBC、NnPEBP、NnPPOA、NnCHS、NnJAZ1和Unknown protein 3)进行一对一验证,发现其中JAZ蛋白与次生代谢物生物碱合成相关,提示NnWRKY40b转录因子可能与茉莉酸(JA)介导的生物碱合成调控密切相关。
Abstract:To explore the potential mechanism of the NnWRKY40 transcription factor in lotus (Nelumbo nucifera Gaertn.) for regulating the synthesis of secondary metabolite alkaloids, a mixed cDNA library from different lotus tissues was constructed, and the proteins interacting with NnWRKY40 were screened. Total RNA of different tissues was extracted from ‘Baihuajian’, and a mixed cDNA library was established. The library capacity was 1.2 × 107 CFU, recombinant rate was 100%, and average length of the inserted fragments was >1 000 bp. NnWRKY40 contains two homologous genes, NnWRKY40a and NnWRKY40b. As NnWRKY40b is reported to play a leading role in transcriptional activation of alkaloid synthesis genes, we used NnWRKY40b to construct the bait vector PGBKT7-NnWRKY40b. In total, 27 proteins interacting with NnWRKY40b were screened from the library using the co-transformation method. These interacting proteins could be divided into three categories: i.e., growth and development and stress response, hormone regulation and secondary metabolism, and unknown proteins. Six representative proteins, including NnUBC, NnPEBP, NnPPOA, NnCHS, NnJAZ1, and unknown protein 3, were selected for one-to-one verification, among which the JAZ protein was associated with alkaloid synthesis, suggesting that the NnWRKY40b transcription factor may be closely related to jasmonic acid (JA)-mediated regulation of alkaloid synthesis in N. nucifera.
-
Keywords:
- Nelumbo nucifera /
- Yeast two-hybrid /
- NnWRKY40 /
- Protein interaction
-
-
表 1 互作蛋白点对点验证所用引物
Table 1 Primers used in one-to-one verification of interacting proteins
引物名
Primer name正向序列(5′–3′)
Sequence of forward primer反向序列(5′–3′)
Sequence of reverse primerpGBKT7-NnWRKY40b CATGGAGGCCGAATTCATGGAGTC
GACTTGGTTGGATACGCAGGTCGACGGATCCTCACCA
TTTCTGCACTGTTGAATGpGADT7-NnJAZ1 CAGATTACGCTCATATGATGTCAA
GAGCGCCGGACCTTGCTTGGGTGGAATTCCTACTGT
GGAGATCGAGCTTGTpGADT7-NnUBC CAGATTACGCTCATATGATGGCGA
ACAGCAATCTACCCTGCTTGGGTGGAATTCTCAGGCA
CCACTTGCATATAGpGADT7-NnCHS CAGATTACGCTCATATGATGGTGA
CCGTGGAAGACATCTGCTTGGGTGGAATTCCTAGGCA
GCGATACTGTGAAGpGADT7-NnPEBP CAGATTACGCTCATATGATGGCGA
GTGACGAGTTTAGGTTGCTTGGGTGGAATTCTTAGGCT
GGGAAAAGTCGGATCpGADT7-NnPPOA CAGATTACGCTCATATGATGGCA
TCGCTTTCTCCCTTGATGCTTGGGTGGAATTCTCACGAA
GCGAACACTATCTTGpGADT7-Unknown protein3 CAGATTACGCTCATATGATGCAT
TCCCTGAGCTTAAAACTTGCTTGGGTGGAATTCTTAGACG
ATATCCGTATCATCTC表 2 NnWRKY40b互作蛋白筛选及其功能预测
Table 2 Screening and functional prediction of NnWRKY40b interacting proteins
分类
Classification蛋白号
Protein ID基因号
Gene ID蛋白名称
Protein name相关蛋白功能预测
Protein function prediction生长发育及
抗逆XP_010271938.1 LOC104607876 枯草杆菌蛋白酶,NnSBT1.7 种皮发育相关 XP_010266914.1 LOC104604316 质体蓝素,NnPC2B 参与光合作用 XP_010279114.1 LOC104613113 泛素结合酶,NnUBC DNA修复,光周期,抗逆胁迫响应,降解生长素,延缓植物衰老,调控ABA信号途径 YP_009093956.1 LOC20834983 ATP合成酶CF1亚基,NnatpE 光合作用,细胞代谢 XP_010265991.1 LOC104603626 类似谷胱甘肽S-转移酶U17, NnGST 抗逆反应,植物修复 XP_010269750.1 LOC104606314 铜转运蛋白5.1,NnCTR5.1 光合作用,呼吸作用,细胞壁代谢,氧化应激反应 XP_010255313.1 LOC104596029 非依赖性蛋白转位酶蛋白,NnTATB 细胞内运输、分泌和囊泡转运 XP_010248208.1 LOC104591115 二磷酸核酮糖羧化酶/加氧酶激活酶,NnRCA 光合作用,叶片衰老,响应非生物胁迫 XP_010270928.1 LOC104607108 半胱氨酸过氧化物氧还蛋白,NnPER1 细胞氧化还原稳态,细胞氧化剂解毒 XP_010269352.1 LOC104606034 60S核糖体蛋白L13e,NnRPL13 翻译、核糖体结构与生物发生 XP_010270872 LOC104607076 ADP-核糖基化因子,NnBLH8 细胞内运输、分泌和囊泡转运 XP_010275748.1 LOC104610704 核糖核酸酶,NnCAF 1 RNA降解 XP_010241640.1 LOC104586181 液泡蛋白分选相关蛋白,NnVPS37-1 盐胁迫响应 XP_010264580.1 LOC104602549 磷脂酰乙醇胺结合蛋白,NnPEBP 植物生长发育,几种信号通路的调节,如MAP激酶通路 XP_010251283.1 LOC104593218 类似Fcf2蛋白,NnFcf 胚成熟,花瓣分化,叶片衰老 XP_010263125.1 LOC104601478 ATP合成酶, NnatpH 光合作用,细胞代谢 XP_010244725.1 LOC104588480 类ACR12蛋白,NnACR12 光合电子传递,冷响应,光响应 激素调控和
次级代谢XP_010258950.1 LOC104598530 泛素蛋白,NnUBQ 蛋白降解,茉莉酸信号途径,细胞周期 XP_010251469.1 LOC104593386 类似TIFY 10A蛋白,NnJAZ1 抑制JA信号传导,响应盐胁迫,
花的发育,茎叶的发育NP_001305084.1 LOC104602160 查尔酮合成酶,NnCHS 类黄酮的生物合成,生长素运输的调节,根系向重力性的调节 XP_010273014.1 LOC104608661 DAHP合成酶,NnDAHP 分支酸合成 ADC92563.1 LOC104588895 多酚氧化酶,NnPPOA 类黄酮、木质素、原花青素生物合成过程 XP_010270953.1 LOC104607120 S-腺苷甲硫氨酸合酶5,NnSAMS 木质素生物合成过程,蛋氨酸代谢过程,冷反应 未知 XP_010261469.1 LOC104600297 未表征蛋白1 未知 XP_010260316.1 LOC104599465 未表征蛋白2 未知 XP_010248518.1 LOC104591415 未表征蛋白3 未知 XP_010276554.1 LOC104611264 未表征蛋白4 未知 -
[1] 陈强,张华,沙玫,刘永静. 一测多评法同时测定荷叶中4种生物碱含量[J]. 福建中医药,2020,51(6):29−32. doi: 10.3969/j.issn.1000-338X.2020.06.011 Chen Q,Zhang H,Sha M,Liu YJ. Simultaneous determination of four alkaloids in nelumbinis folium by quantitative analysis of multi-components by single marker[J]. Fujian Journal of Traditional Chinese Medicine,2020,51 (6):29−32. doi: 10.3969/j.issn.1000-338X.2020.06.011
[2] Wan Y,Xia J,Xu JF,Chen L,Yang Y,et al. Nuciferine,an active ingredient derived from lotus leaf,lights up the way for the potential treatment of obesity and obesity-related diseases[J]. Pharmacol Res,2022,175:106002. doi: 10.1016/j.phrs.2021.106002
[3] Abdallah BM,Ali EM. Green synthesis of silver nanoparticles using the Lotus lalambensis aqueous leaf extract and their anti-candidal activity against oral candidiasis[J]. ACS Omega,2021,6 (12):8151−8162. doi: 10.1021/acsomega.0c06009
[4] Tong YL,Li ZW,Wu YK,Zhu SL,Lu KK,He Z. Lotus leaf extract inhibits ER- breast cancer cell migration and metastasis[J]. Nutr Metab,2021,18 (1):20. doi: 10.1186/s12986-021-00549-0
[5] Van der Fits L,Memelink J. ORCA3,a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism[J]. Science,2000,289 (5477):295−297. doi: 10.1126/science.289.5477.295
[6] Kato N,Dubouzet E,Kokabu Y,Yoshida S,Taniguchi Y,et al. Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica[J]. Plant Cell Physiol,2007,48 (1):8−18. doi: 10.1093/pcp/pcl041
[7] Suttipanta N,Pattanaik S,Kulshrestha M,Patra B,Singh SK,Yuan L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus[J]. Plant Physiol,2011,157 (4):2081−2093. doi: 10.1104/pp.111.181834
[8] Agarwal P,Pathak S,Lakhwani D,Gupta P,Asif MH,Trivedi PK. Comparative analysis of transcription factor gene families from Papaver somniferum:identification of regulatory factors involved in benzylisoquinoline alkaloid biosynthesis[J]. Protoplasma,2016,253 (3):857−871. doi: 10.1007/s00709-015-0848-8
[9] Zhou ML,Memelink J. Jasmonate-responsive transcription factors regulating plant secondary metabolism[J]. Biotechnol Adv,2016,34 (4):441−449. doi: 10.1016/j.biotechadv.2016.02.004
[10] Tripathi S,Sangwan RS,Mishra B,Jadaun JS,Sangwan NS. Berry transcriptome:insights into a novel resource to understand development dependent secondary metabolism in Withania somnifera(Ashwagandha)[J]. Physiol Plant,2020,168 (1):148−173. doi: 10.1111/ppl.12943
[11] Hao XL,Xie CH,Ruan QY,Zhang XC,Wu C,et al. The transcription factor OpWRKY2 positively regulates the biosynthesis of the anticancer drug camptothecin in Ophiorrhiza pumila[J]. Hortic Res,2021,8 (1):7. doi: 10.1038/s41438-020-00437-3
[12] Eulgem T,Rushton PJ,Robatzek S,Somssich IE. The WRKY superfamily of plant transcription factors[J]. Trends Plant Sci,2000,5 (5):199−206. doi: 10.1016/S1360-1385(00)01600-9
[13] Ishiguro S,Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein,SPF1,that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato[J]. Mol Gen Genet,1994,244 (6):563−571. doi: 10.1007/BF00282746
[14] 向小华,吴新儒,晁江涛,杨明磊,杨帆,等. 普通烟草WRKY基因家族的鉴定及表达分析[J]. 遗传,2016,38(9):840−856. doi: 10.16288/j.yczz.16-016 Xiang XH,Wu XR,Chao JT,Yang ML,Yang F,et al. Genome-wide identification and expression analysis of the WRKY gene family in common tobacco (Nicotiana tabacum L. )[J]. Hereditas,2016,38 (9):840−856. doi: 10.16288/j.yczz.16-016
[15] Cormack RS,Eulgem T,Rushton PJ,Köchner P,Hahlbrock K,Somssich IE. Leucine zipper-containing WRKY proteins widen the spectrum of immediate early elicitor-induced WRKY transcription factors in parsley[J]. BBA-Gene Struct Expr,2002,1576 (1-2):92−100. doi: 10.1016/S0167-4781(02)00298-1
[16] 郑超,郑二松,王栩鸣,李冬月,杨勇,等. 水稻WRKY转录调控因子研究进展[J]. 生物技术通讯,2018,29(2):286−294. doi: 10.3969/j.issn.1009-0002.2018.02.026 Zheng C,Zheng ES,Wang XM,Li DY,Yang Y,et al. Research progress on rice WRKY transcription factors[J]. Letters in Biotechnology,2018,29 (2):286−294. doi: 10.3969/j.issn.1009-0002.2018.02.026
[17] 杨致荣,王兴春,薛金爱,孟令芝,李润植. 药用植物长春花WRKY转录因子的鉴定及表达谱分析[J]. 生物工程学报,2013,29(6):785−802. doi: 10.13345/j.cjb.2013.06.006 Yang ZR,Wang XC,Xue JA,Meng LZ,Li RZ. Identification and expression analysis of WRKY transcription factors in medicinal plant Catharanthus roseus[J]. Chinese Journal of Biotechnology,2013,29 (6):785−802. doi: 10.13345/j.cjb.2013.06.006
[18] Yamada Y,Nishida S,Shitan N,Sato F. Genome-wide profiling of WRKY genes involved in benzylisoquinoline alkaloid biosynthesis in California Poppy (Eschscholzia californica)[J]. Front Plant Sci,2021,2:699326.
[19] Wei HW,Chen SY,Niyitanga S,Liu T,Qi JM,Zhang LW. Genome-wide identification and expression analysis response to GA3 stresses of WRKY gene family in seed hemp (Cannabis sativa L. )[J]. Gene,2022,822:146290. doi: 10.1016/j.gene.2022.146290
[20] Mishra S,Triptahi V,Singh S,Phukan UJ,Gupta MM,et al. Wound induced tanscriptional regulation of benzylisoquinoline pathway and characterization of wound inducible PsWRKY transcription factor from Papaver somniferum[J]. PLoS One,2013,8 (1):e52784. doi: 10.1371/journal.pone.0052784
[21] He J,Bouwmeester HJ,Dicke M,Kappers IF. Transcriptional and metabolite analysis reveal a shift in direct and indirect defences in response to spider-mite infestation in cucumber (Cucumis sativus)[J]. Plant Mol Biol,2020,103 (4-5):489−505. doi: 10.1007/s11103-020-01005-y
[22] 代红洋,柏旭,李晓岗,张兴开,罗霖,等. 植物激素在三萜类化合物生物合成中的作用及调控机制研究进展[J]. 中草药,2021,52(20):6391−6402. Dai HY,Bai X,Li XG,Zhang XK,Luo L,et al. Research progress on roles of phytohormone in biosynthesis of triterpenoids and their regulatory mechanisms[J]. Chinese Traditional and Herbal Drugs,2021,52 (20):6391−6402.
[23] Wasternack C,Song SS. Jasmonates:biosynthesis,metabolism,and signaling by proteins activating and repressing transcription[J]. J Exp Bot,2017,68 (6):1303−1321.
[24] Yang J,Duan GH,Li CQ,Liu L,Han GY,et al. The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses[J]. Front Plant Sci,2019,10:1349. doi: 10.3389/fpls.2019.01349
[25] Ming R,VanBuren R,Liu YL,Yang M,Han YP,et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn. )[J]. Genome Biol,2013,14 (5):R41. doi: 10.1186/gb-2013-14-5-r41
[26] Zhang Y,Rahmani RS,Yang XY,Chen JM,Shi T. Integrative expression network analysis of microRNA and gene isoforms in sacred lotus[J]. BMC Genomics,2020,21 (1):429. doi: 10.1186/s12864-020-06853-y
[27] Li J,Xiong YC,Li Y,Ye SQ,Yin Q,et al. Comprehensive analysis and functional studies of WRKY transcription factors in Nelumbo nucifera[J]. Int J Mol Sci,2019,20 (20):5006. doi: 10.3390/ijms20205006
[28] Ferrer JL,Austin MB,Stewart C Jr,Noel JP. Structure and function of enzymes involved in the biosynthesis of phenylpropanoids[J]. Plant Physiol Biochem,2008,46 (3):356−370. doi: 10.1016/j.plaphy.2007.12.009
[29] Ren GH,Wang BJ,Zhu XD,Mu Q,Wang C,et al. Cloning,expression,and characterization of miR058 and its target PPO during the development of grapevine berry stone[J]. Gene,2014,548 (2):166−173. doi: 10.1016/j.gene.2014.07.021
[30] 王馨雨,杨绿竹,王婷,王蓉蓉,刘洁,等. 植物多酚氧化酶的生理功能、分离纯化及酶促褐变控制的研究进展[J]. 食品科学,2020,41(9):222−237. doi: 10.7506/spkx1002-6630-20190411-145 Wang XY,Yang LZ,Wang T,Wang RR,Liu J,et al. Recent progress toward understanding the physiological function,purification,and enzymatic browning control of plant polyphenol oxidases[J]. Food Science,2020,41 (9):222−237. doi: 10.7506/spkx1002-6630-20190411-145
[31] 田娇,刘园,房敏峰. 外源茉莉酸类激素对药用植物次生代谢的影响研究[J]. 天然产物研究与开发,2015,27(1):185−190. doi: 10.16333/j.1001-6880.2015.01.037 Tian J,Liu Y,Fang MF. Review on the influence of exogenous jasmonates on medicinal plant secondary metabolism[J]. Natural Product Research and Development,2015,27 (1):185−190. doi: 10.16333/j.1001-6880.2015.01.037
[32] 王金利,史胜青,贾利强,江泽平. 植物泛素结合酶E2功能研究进展[J]. 生物技术通报,2010(4):7−10. doi: 10.13560/j.cnki.biotech.bull.1985.2010.04.002 Wang JL,Shi SQ,Jia LQ,Jiang ZP. Progress on functions of ubiquitin-conjugating enzyme (E2) in plants[J]. Biotechnology Bulletin,2010 (4):7−10. doi: 10.13560/j.cnki.biotech.bull.1985.2010.04.002
[33] 李兴芬,苗雅慧,孙永江,张孟娟,张凌云. 青杄PwPEBP基因及其启动子序列的克隆与表达分析[J]. 北京林业大学学报,2019,41(4):8−20. Li XF,Miao YH,Sun YJ,Zhang MJ,Zhang LY. Cloning and expression analysis of PwPEBP gene and promoter sequence in Picea wilsonii[J]. Journal of Beijing Forestry University,2019,41 (4):8−20.
[34] 王寻,高凝,张富军,韩月彭,王小非,等. 苹果磷脂酰乙醇胺结合蛋白PEBP家族基因的鉴定与比较分析[J]. 植物生理学报,2021,57(10):1996−2010. doi: 10.13592/j.cnki.ppj.2020.0395 Wang X,Gao N,Zhang FJ,Han YP,Wang XF,et al. Identification and comparative analysis of phosphatidyl ethanolamine binding protein (PEBP) family gene in apple[J]. Plant Physiology Journal,2021,57 (10):1996−2010. doi: 10.13592/j.cnki.ppj.2020.0395
[35] 祝一文,车永梅,赵方贵,朱丹,刘新. 碱胁迫下H2S参与活性氧代谢和水稻幼苗生长的调控[J]. 农业生物技术学报,2018,26(7):1124−1131. Zhu YW,Che YM,Zhao FG,Zhu D,Liu X. H2S functions in growth regulation in rice (Oryza sativa) seedling and metabolism modulating of reactive oxygen under alkaline stress[J]. Journal of Agricultural Biotechnology,2018,26 (7):1124−1131.
[36] 张金梅,白雪,李玥莹,张颖. WRKY响应植物逆境的“角色”[J]. 安徽农业科学,2020,48(12):5−8. doi: 10.3969/j.issn.0517-6611.2020.12.002 Zhang JM,Bai X,Li YY,Zhang Y. WRKY’s “Role” in response to plant adversity[J]. Journal of Anhui Agricultural Sciences,2020,48 (12):5−8. doi: 10.3969/j.issn.0517-6611.2020.12.002
[37] 魏昕,刘雨恒,刘宇阳,殷晓浦,谢恬,等. 植物JAZ蛋白家族研究进展[J]. 植物生理学报,2021,57(5):1039−1046. doi: 10.13592/j.cnki.ppj.2020.0532 Wei X,Liu YH,Liu YY,Yin XP,Xie T,et al. Advances of JAZ family in plants[J]. Plant Physiology Journal,2021,57 (5):1039−1046. doi: 10.13592/j.cnki.ppj.2020.0532
[38] Chini A,Fonseca S,Fernández G,Adie B,Chico JM,et al. The JAZ family of repressors is the missing link in jasmonate signalling[J]. Nature,2007,448 (7154):666−671. doi: 10.1038/nature06006
[39] Thines B,Katsir L,Melotto M,Niu YJ,Mandaokar A,et al. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling[J]. Nature,2007,448 (7154):661−665. doi: 10.1038/nature05960
[40] Chen XZ,Li JR,Liu YT,Wu DD,Huang HL,et al. PatSWC4,a methyl jasmonate-responsive MYB (v-myb avian myeloblastosis viral oncogene homolog)-related transcription factor,positively regulates patchoulol biosynthesis in Pogostemon cablin[J]. Ind Crops Prod,2020,154:112672. doi: 10.1016/j.indcrop.2020.112672
-
期刊类型引用(1)
1. 惠生娟,葛丽萍,王子瑜,张玉胜,苏云婷,孙岩,李润植. 续随子MYB基因家族的鉴定及ElMYB114在油脂合成中的功能分析. 植物科学学报. 2025(01): 92-101 .  本站查看
本站查看
其他类型引用(0)



 下载:
下载: